Search Count: 459
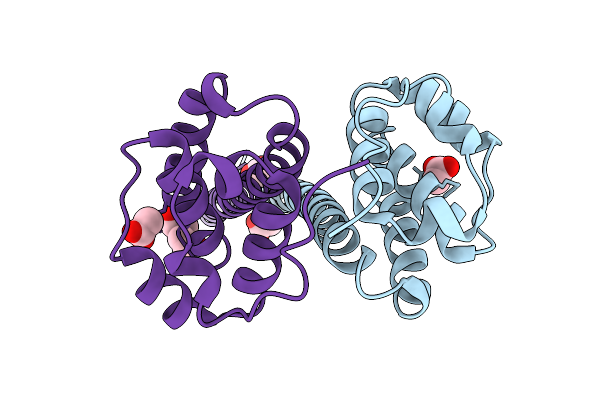 |
Outer Membrane Lipoprotein Qseg Of Salmonella Enterica Serovar Typhimurium Sl1344
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SIGNALING PROTEIN Ligands: ACT, PEG |
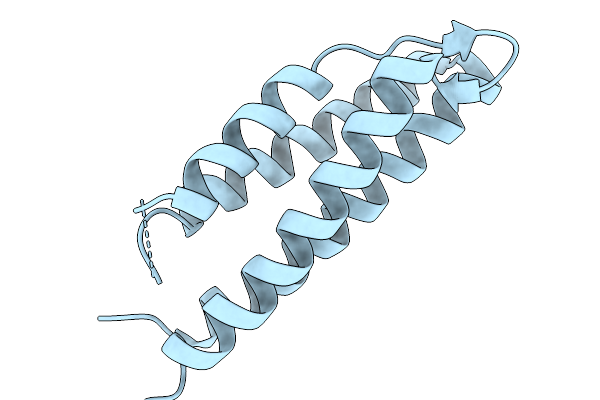 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
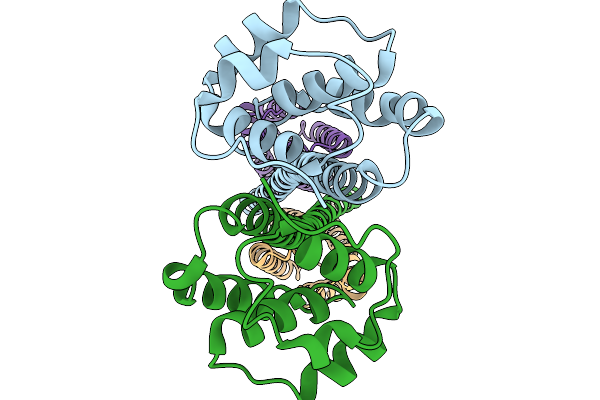 |
Structure Of Outer Membrane Lipoprotein Qseg And Histidine Kinase Qsee Complex
Organism: Salmonella enterica subsp. enterica serovar typhimurium str. sl1344
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: SIGNALING PROTEIN |
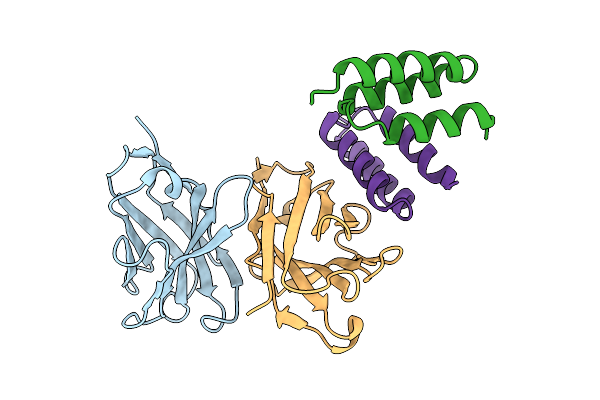 |
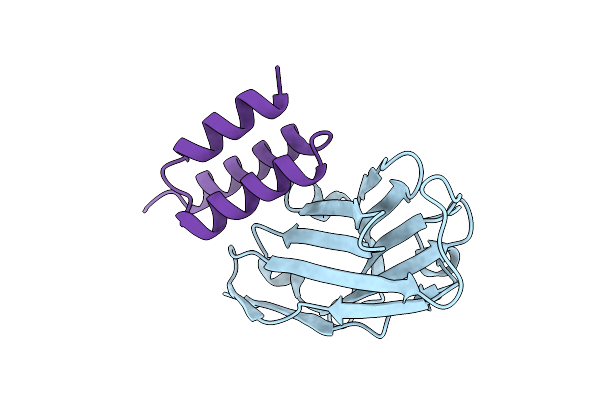
Crystal Structure Of S. Aureus Protein A Bound To A Human Single-Domain Antibody
Organism: Homo sapiens, Staphylococcus aureus (strain nctc 8325 / ps 47)
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
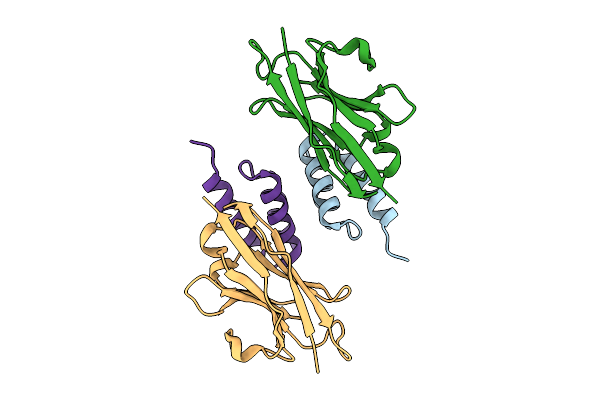
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Camelidae, Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Crystal Structure Of S. Aureus Protein A Bound To A Camelid Single-Domain Antibody
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Camelus dromedarius
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: IMMUNE SYSTEM |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
 |
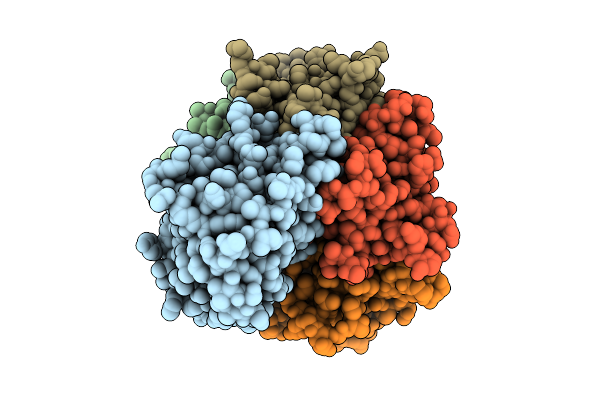
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
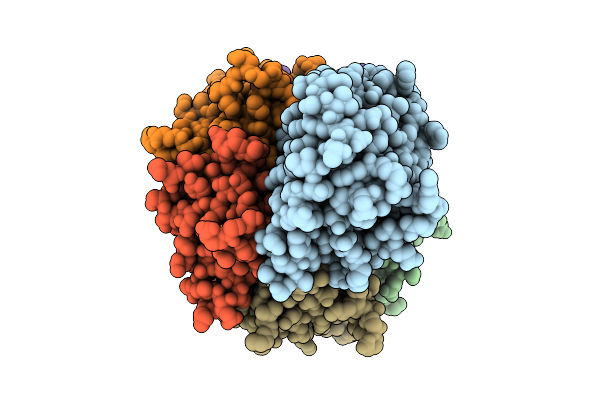 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Campylobacter jejuni subsp. jejuni serotype o:23/36 (strain 81-176)
Method: ELECTRON MICROSCOPY Release Date: 2025-10-15 Classification: STRUCTURAL PROTEIN |
 |
Organism: Escherichia coli (strain k12), Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: TOXIN |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: ANTITOXIN Ligands: IPA |
 |
Organism: Bacteroides fragilis nctc 9343
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: STRUCTURAL PROTEIN |
 |
Structure Of Cargo Complex (Btpea-Btaeb-Btapc) Bound To The Vgrg Spike From The Type Vi Secretion System
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN |
 |
Structure Of The Btpea Effector And Btaeb Effector Bound To The Vgrg Spike From The Type Vi Secretion System
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN |
 |
Crystal Structure Of The Type Vi Secretion System Effector-Immunity Complex Btaeb Ctd-Btaib From Bacteroides Fragilis
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: GOL, CA |
 |
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: PEG |
 |
Crystal Structure Of The Type Vi Secretion System Effector-Immunity Complex Btpea Ctd-Btpia From Bacteroides Fragilis.
Organism: Bacteroides fragilis
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: ANTIMICROBIAL PROTEIN Ligands: GOL |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: ANTIVIRAL PROTEIN |

