Search Count: 59
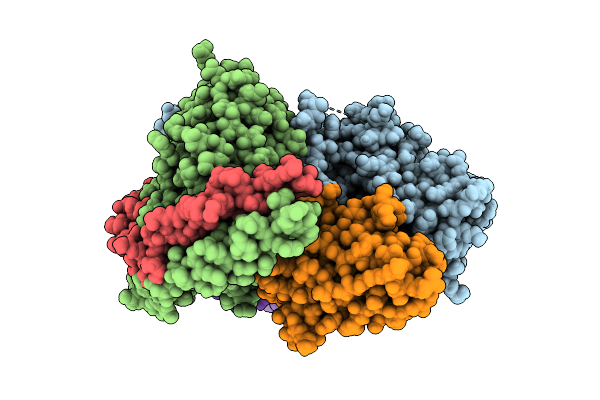 |
Cryo-Em Structure Of The Proton-Sensing Gpcr (Gpr4)-Gs Protein Complex At Ph 6.5
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN Ligands: CLR, A1L35 |
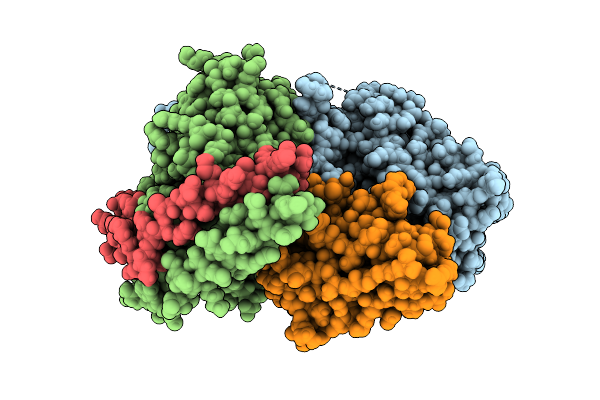 |
Cryo-Em Structure Of The Proton-Sensing Gpcr (Gpr4)-Gq Protein Complex At Ph 7.4
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN Ligands: CLR, A1L35 |
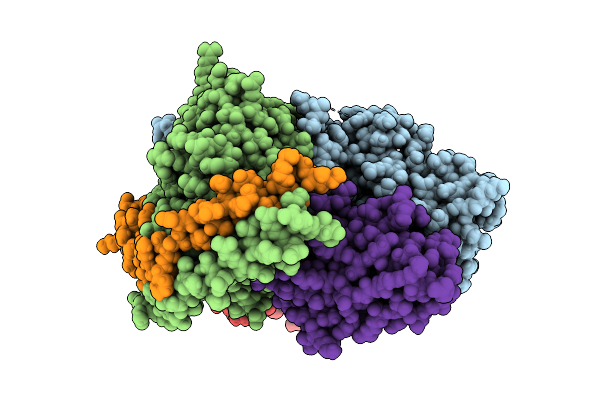 |
Cryo-Em Structure Of The Proton-Sensing Gpcr (Gpr4)-Gs Protein Complex At Ph 7.4
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-07-09 Classification: SIGNALING PROTEIN Ligands: A1L35, CLR |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: NUCLEAR PROTEIN |
 |
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-07-02 Classification: NUCLEAR PROTEIN Ligands: ZN |
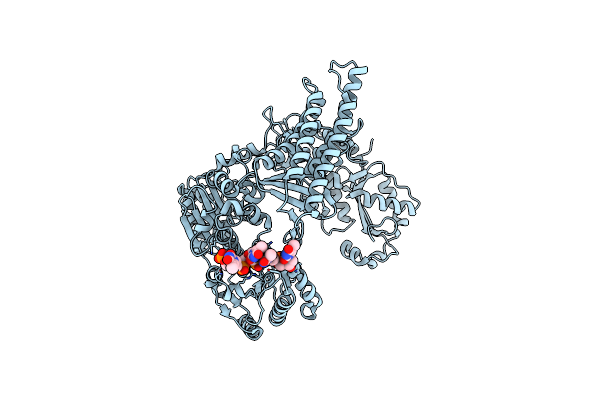 |
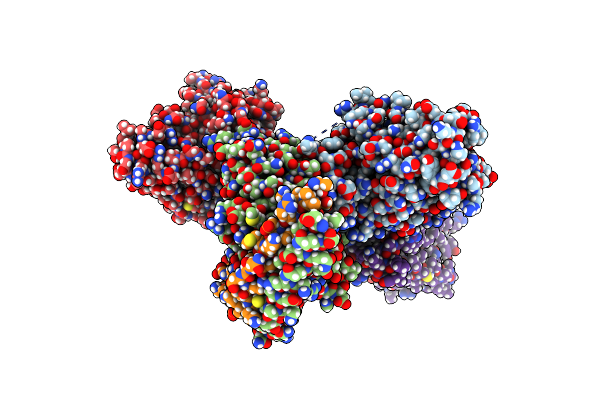
Crystal Structure Of Staphylococcus Aureus Ding Protein In Complex With Ssdna
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: HYDROLASE/DNA |
 |
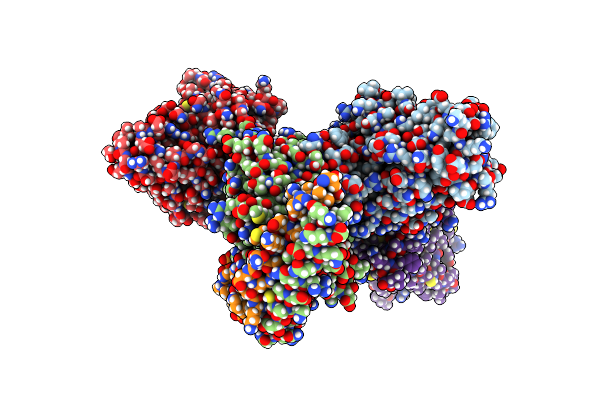
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: SIGNALING PROTEIN Ligands: CLR, A1L4H |
 |
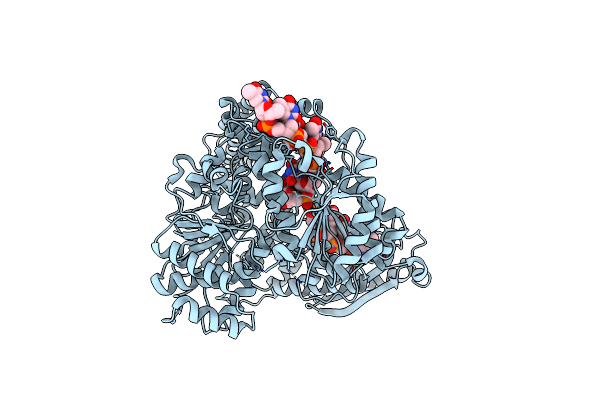
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: SIGNALING PROTEIN Ligands: CLR, ZIT |
 |
Crystal Structure Of Staphylococcus Aureus Ding Protein In Complex With Ssdna And Ca2+
Organism: Staphylococcus aureus (strain nctc 8325 / ps 47), Chemical production metagenome
Method: X-RAY DIFFRACTION Resolution:3.21 Å Release Date: 2025-05-21 Classification: HYDROLASE/DNA Ligands: CA |
 |
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN |
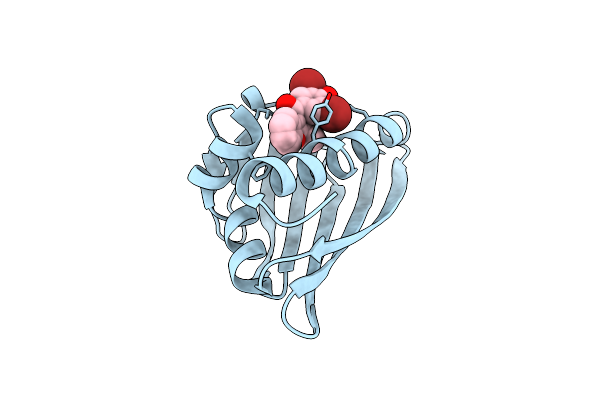 |
Benzbromarone Interferes With The Interaction Between Hsp90 And Aha1 By Interacting With Both Of Them
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-04-23 Classification: STRUCTURAL PROTEIN Ligands: R75 |
 |
Crystal Structure Of Integrin Beta-3 Tail Bound To The Ferm-Folded Talin Head Domain With A D397R Mutation
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-23 Classification: CELL ADHESION Ligands: SO4 |
 |
Crystal Structure Of Integrin Beta-2 Tail Bound To The Ferm-Folded Talin Head Domain With A D397R Mutation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2025-04-23 Classification: CELL ADHESION |
 |
Crystal Structure Of Integrin Beta-2 Tail Bound To The Ferm-Folded Talin Head Domain With A Y270E Mutation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2024-10-30 Classification: CELL ADHESION |
 |
Crystal Structure Of Integrin Beta-6 Tail Bound To The Ferm-Folded Talin Head Domain
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2024-10-16 Classification: CELL ADHESION |
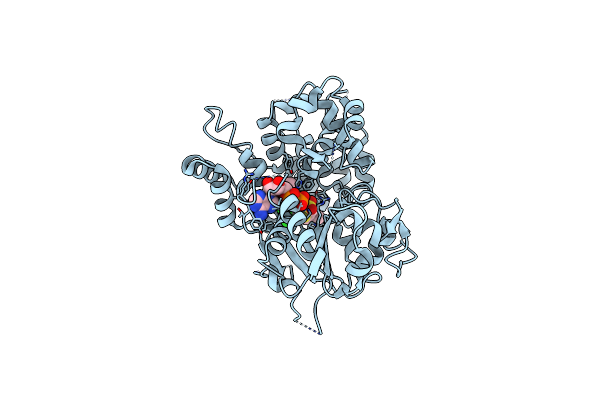 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.32 Å Release Date: 2024-10-02 Classification: IMMUNE SYSTEM Ligands: A1D79, ADP |
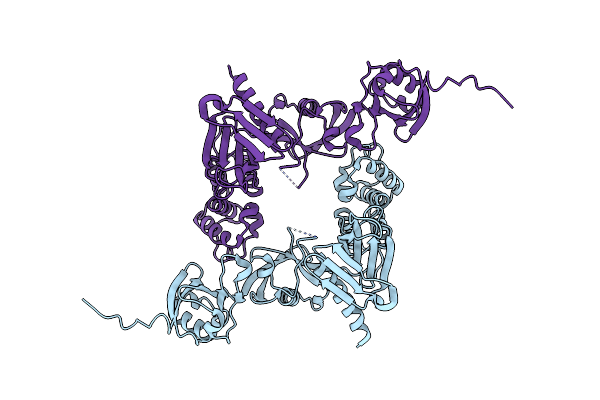 |
Crystal Structure Of Integrin Beta-2 Tail Bound To The Ferm-Folded Talin Head Domain
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-07-03 Classification: CELL ADHESION |
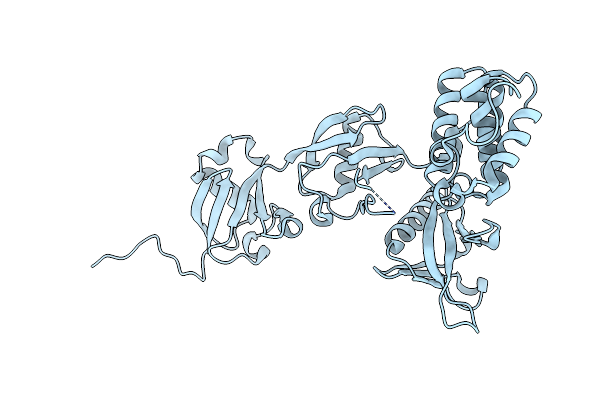 |
Crystal Structure Of Integrin Beta-2 Tail Bound To The Ferm-Folded Talin Head Domain With A K306Q Mutation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2024-07-03 Classification: CELL ADHESION |
 |
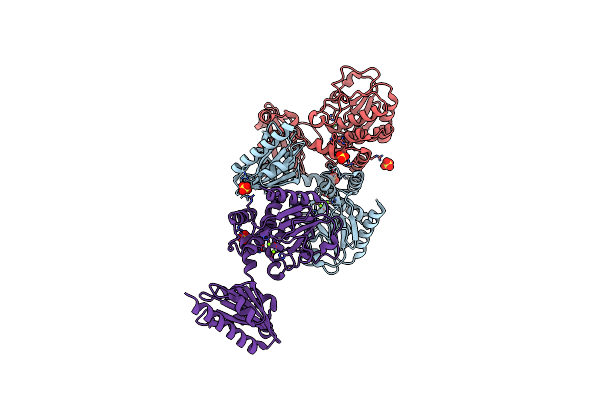
Crystal Structure Of Integrin Beta-2 Tail Bound To The Ferm-Folded Talin Head Domain With E269A/Y270A/K306Q Triple Mutation
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.77 Å Release Date: 2024-07-03 Classification: CELL ADHESION |
 |
Crystal Structure Of Deinococcus Radiodurans Recj-Like Protein In Complex With Mg2+
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-03-20 Classification: HYDROLASE Ligands: MG, SO4 |

