Search Count: 334
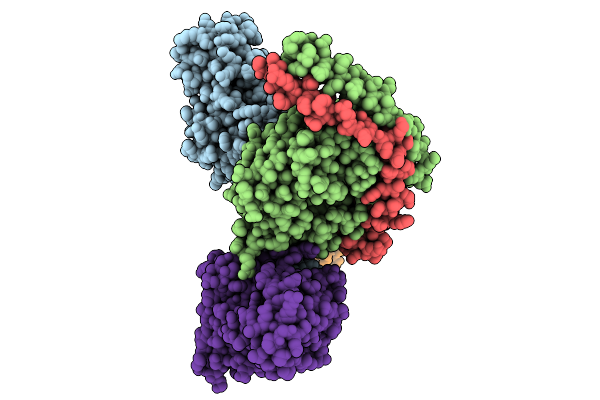 |
Consensus Olfactory Receptor Consor6 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.78 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJQ, CLR |
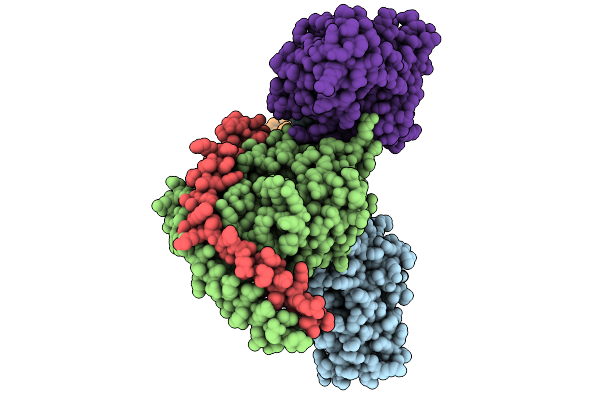 |
Consensus Olfactory Receptor Consor6 Bound To Alpha-Hexyl Cinnamaldehyde And In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.62 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: CLR, A1EJG |
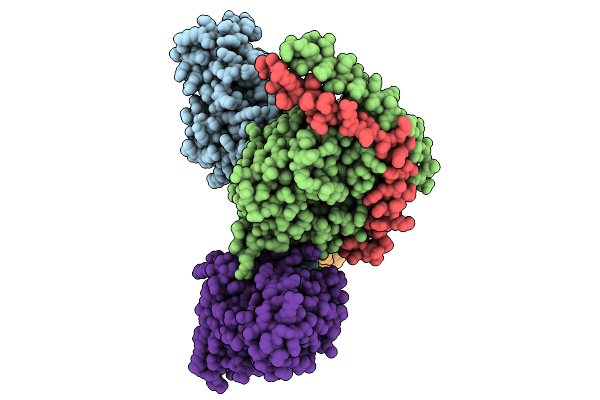 |
Consensus Olfactory Receptor Consor6 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:2.83 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: CLR, BZQ |
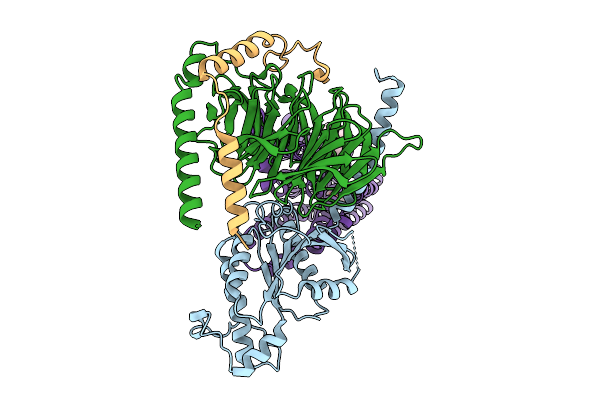 |
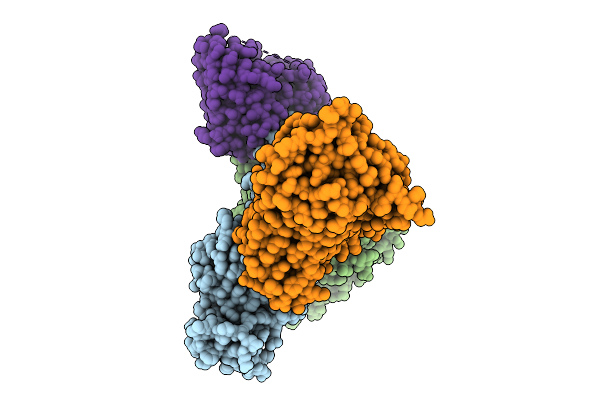
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.17 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN Ligands: A1EJQ, CLR |
 |
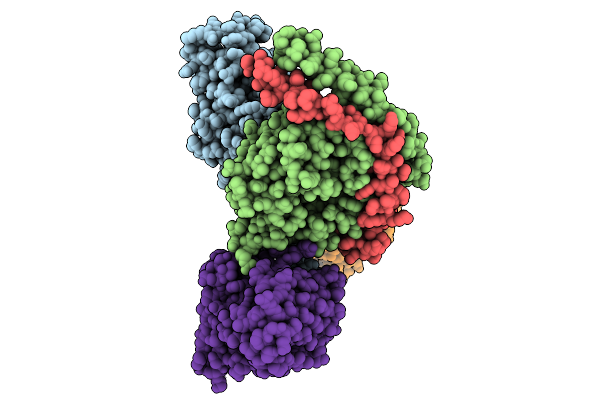
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Mus musculoides, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:2.54 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: OYA |
 |
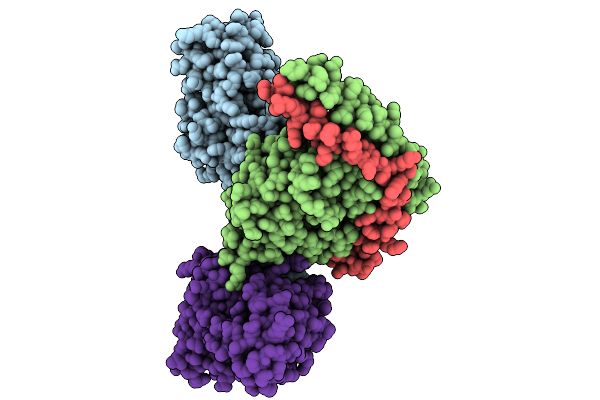
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJH |
 |
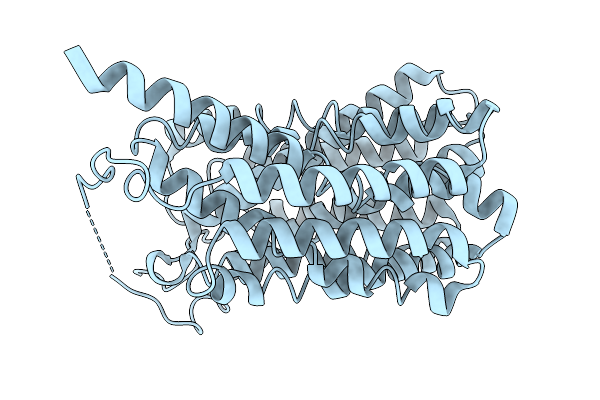
Consensus Olfactory Receptor Bmor6A2 In Complex With Mini-Golf Trimeric Protein
Organism: Homo sapiens, Synthetic construct, Mus musculoides
Method: ELECTRON MICROSCOPY Resolution:3.33 Å Release Date: 2026-01-28 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: A1EJH |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.07 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.16 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: GLP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.47 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.12 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.89 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.89 Å Release Date: 2026-01-07 Classification: TRANSPORT PROTEIN Ligands: PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2025-12-17 Classification: IMMUNE SYSTEM |
 |
Organism: Tetrahymena thermophila
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: GOL |
 |
Organism: Tetrahymena thermophila
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: MG, GOL |
 |
Organism: Tetrahymena thermophila
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: WO4, MG, GOL |
 |
Organism: Tetrahymena thermophila
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-11-12 Classification: HYDROLASE Ligands: CA, GOL |

