Search Count: 40
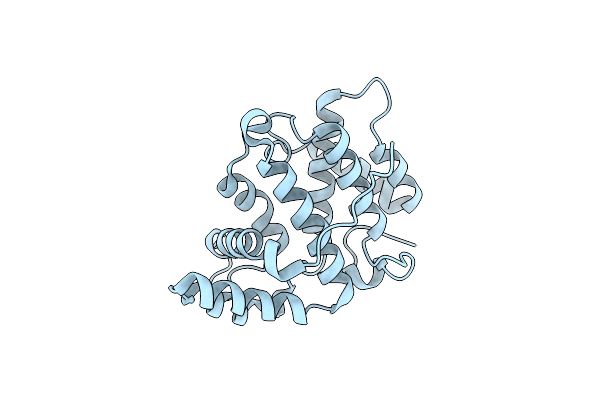 |
Crystal Structure Of The Rhogap Domain Of Rgd1P At 2.19 Angstroms Resolution
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-01-24 Classification: CELL CYCLE |
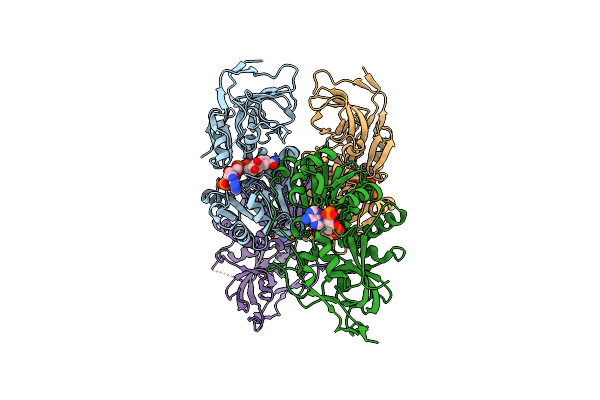 |
Structure Of The Binary Complex Of A Zingiber Officinale Double Bond Reductase In Complex With Nadp Monoclinic Crystal Form
Organism: Zingiber officinale
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAP |
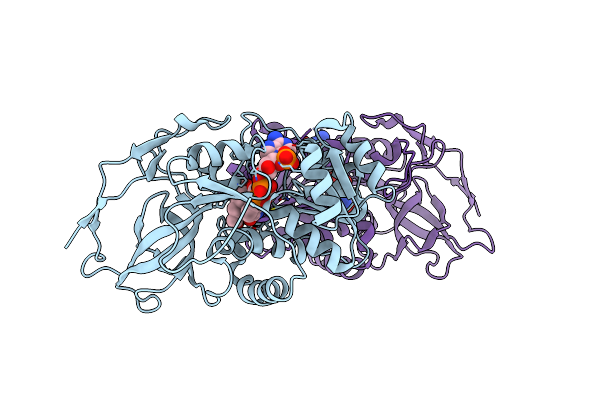 |
Structure Of The Ternary Complex Of A Zingiber Officinale Double Bond Reductase In Complex With Nadp And Coniferyl Aldehyde
Organism: Zingiber officinale
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAP, CIY |
 |
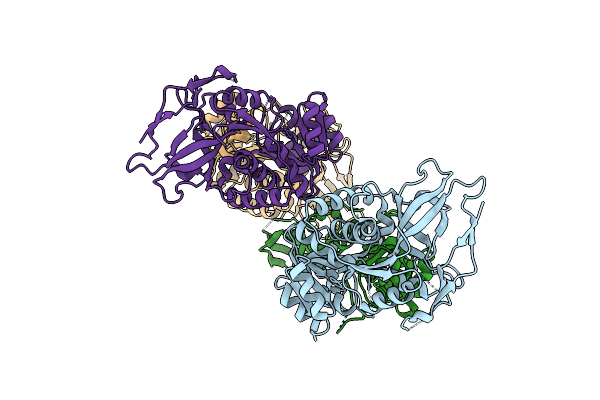
Structure Of The Binary Complex Of A Zingiber Officinale Double Bond Reductase In Complex With Nadp
Organism: Zingiber officinale
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-11-05 Classification: PLANT PROTEIN Ligands: NAP |
 |
Organism: Zingiber officinale
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2014-09-10 Classification: PLANT PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.00 Å Release Date: 2013-12-11 Classification: LYASE/LYASE INHIBITOR Ligands: ZN, SBW, HGB, GOL, HG |
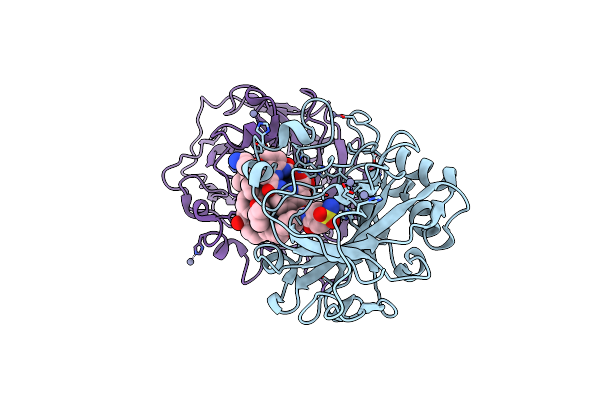 |
Crystal Structure Of Human Carbonic Anhydrase Ii In Complex With A Quinoline Oligoamide Foldamer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2013-10-23 Classification: Lyase/lyase Inhibitor Ligands: Q4I, ZN |
 |
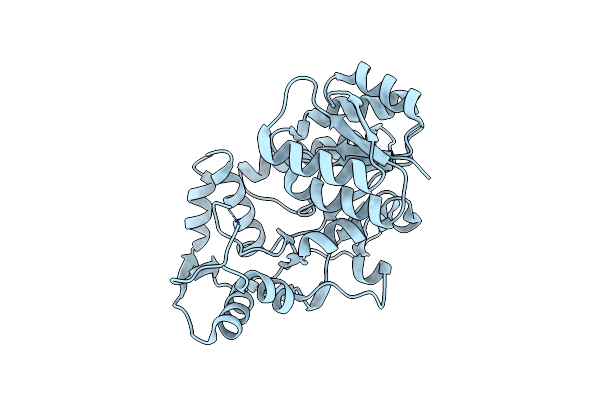
Ternary Complex Structure Of Leucoanthocyanidin Reductase From Vitis Vinifera
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2010-02-23 Classification: OXIDOREDUCTASE Ligands: NAP, KXN |
 |
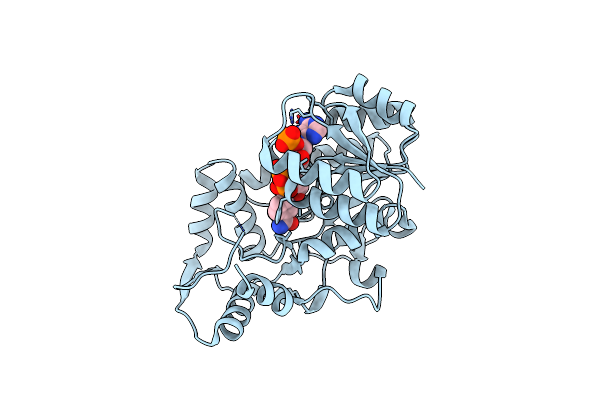
Structure Of The Apo Form Of Leucoanthocyanidin Reductase From Vitis Vinifera
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2010-02-23 Classification: OXIDOREDUCTASE |
 |
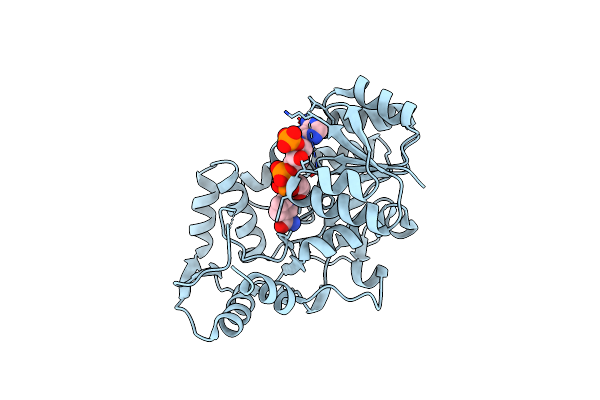
Structure Of The Binary Complex Leucoanthocyanidin Reductase - Nadph From Vitis Vinifera
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2010-02-23 Classification: OXIDOREDUCTASE Ligands: NDP, ACY |
 |
Structure Of The Binary Complex Leucoanthocyanidin Reductase-Nadph From Vitis Vinifera
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2010-02-23 Classification: OXIDOREDUCTASE Ligands: NDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-02-09 Classification: METAL BINDING PROTEIN Ligands: CD, SO4 |
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:3.17 Å Release Date: 2009-09-01 Classification: OXIDOREDUCTASE Ligands: CL |
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2008-11-18 Classification: OXIDOREDUCTASE Ligands: CL |
 |
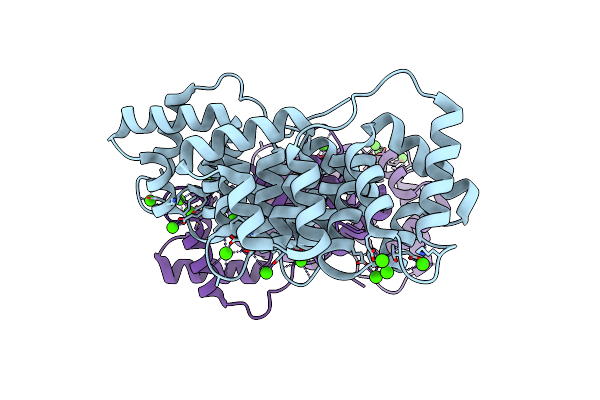
Binding Of Two Substrate Analogue Molecules To Dihydroflavonol 4-Reductase Alters The Functional Geometry Of The Catalytic Site
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-10-21 Classification: OXIDOREDUCTASE Ligands: NAP, QUE |
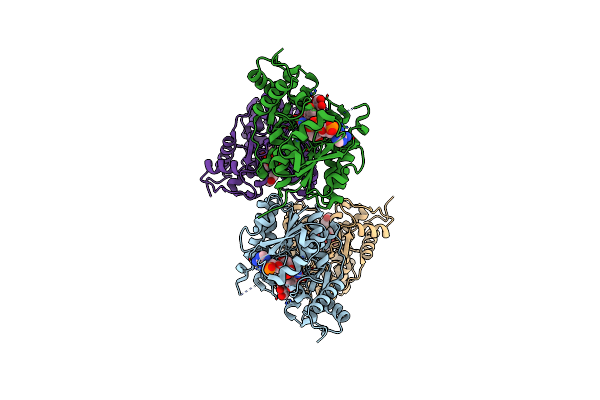 |
Binding Of Two Substrate Analogue Molecules To Dihydroflavonol 4-Reductase Alters The Functional Geometry Of The Catalytic Site
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2008-02-19 Classification: OXIDOREDUCTASE Ligands: NAP, MYC |
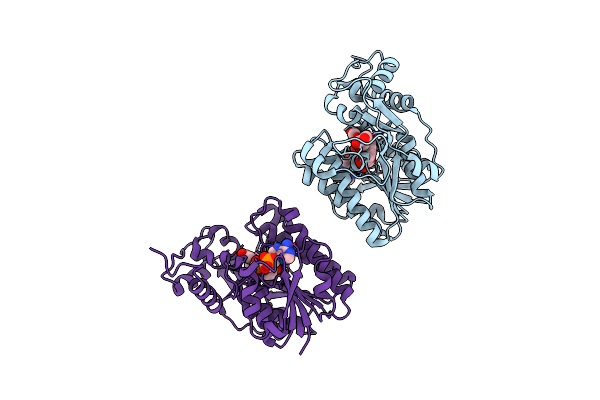 |
Binding Of Two Substrate Analogue Molecules To Dihydroflavonol-4-Reductase Alters The Functional Geometry Of The Catalytic Site
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2007-11-13 Classification: OXIDOREDUCTASE Ligands: NAP, ERD |
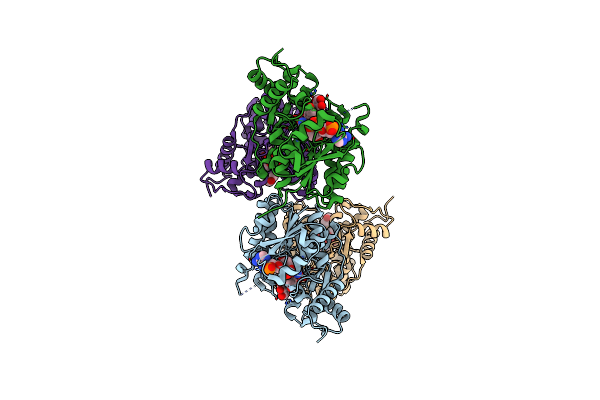 |
Binding Of Two Substrate Analogue Molecules To Dihydroflavonol-4-Reductase Alters The Functional Geometry Of The Catalytic Site
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2007-09-11 Classification: OXIDOREDUCTASE Ligands: NAP, MYC |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.76 Å Release Date: 2007-06-19 Classification: BLOOD CLOTTING Ligands: CA |
 |
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2007-06-19 Classification: BLOOD CLOTTING Ligands: CA |

