Search Count: 26
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, MG, A1BEI |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2025-01-22 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: GDP, A1BEJ, MG |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-01-31 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: GOL, SF4, MPD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: SF4, MPD |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2022-06-29 Classification: REPLICATION Ligands: SF4, MPD |
 |
Model Of Haliangium Ochraceum Encapsulin From Icosahedral Single Particle Reconstruction
Organism: Haliangium ochraceum (strain dsm 14365 / jcm 11303 / smp-2)
Method: ELECTRON MICROSCOPY Release Date: 2022-03-09 Classification: VIRUS LIKE PARTICLE |
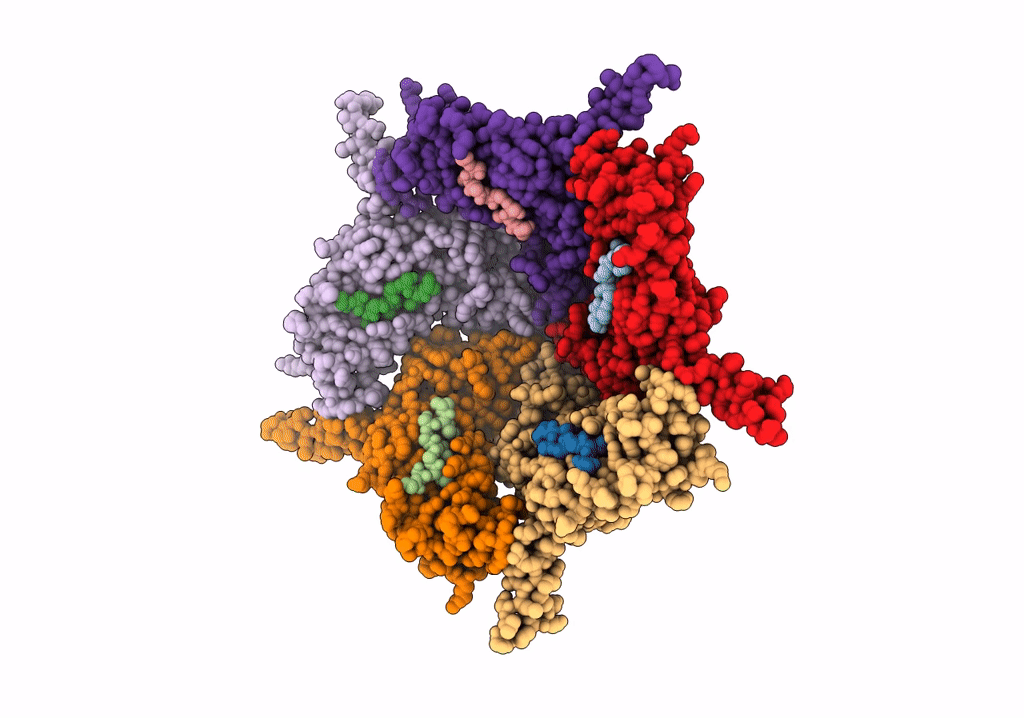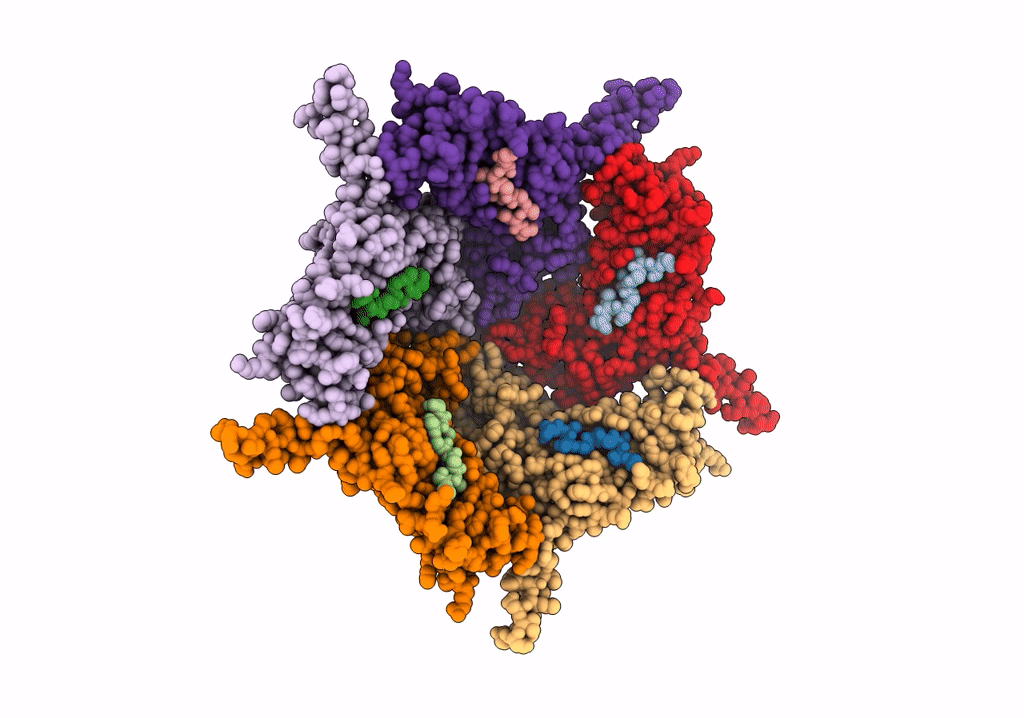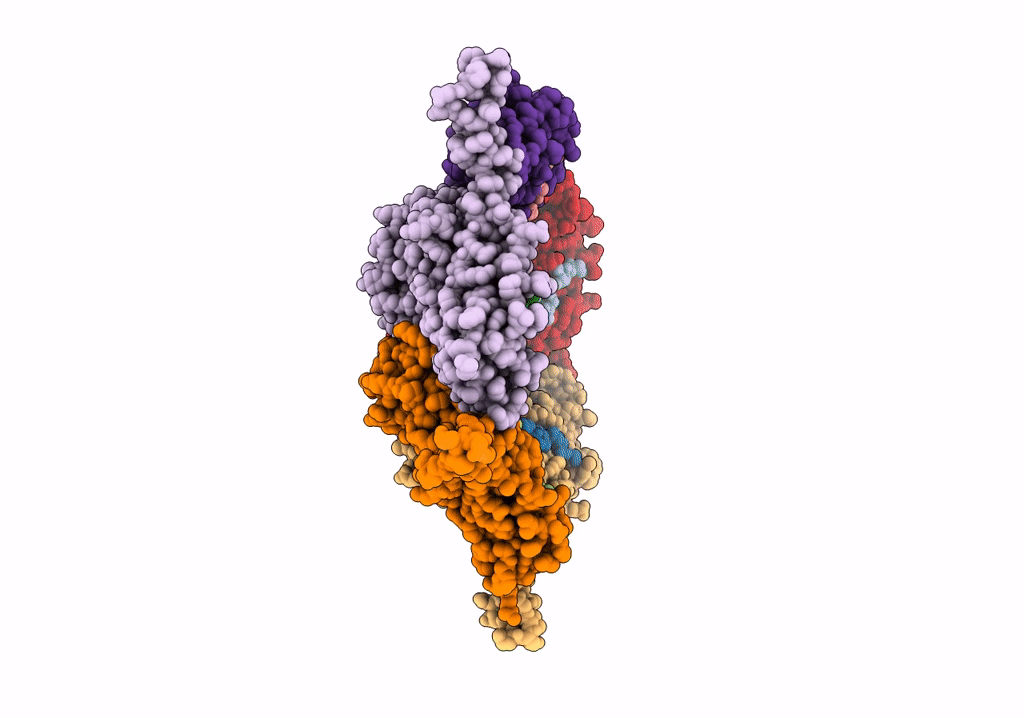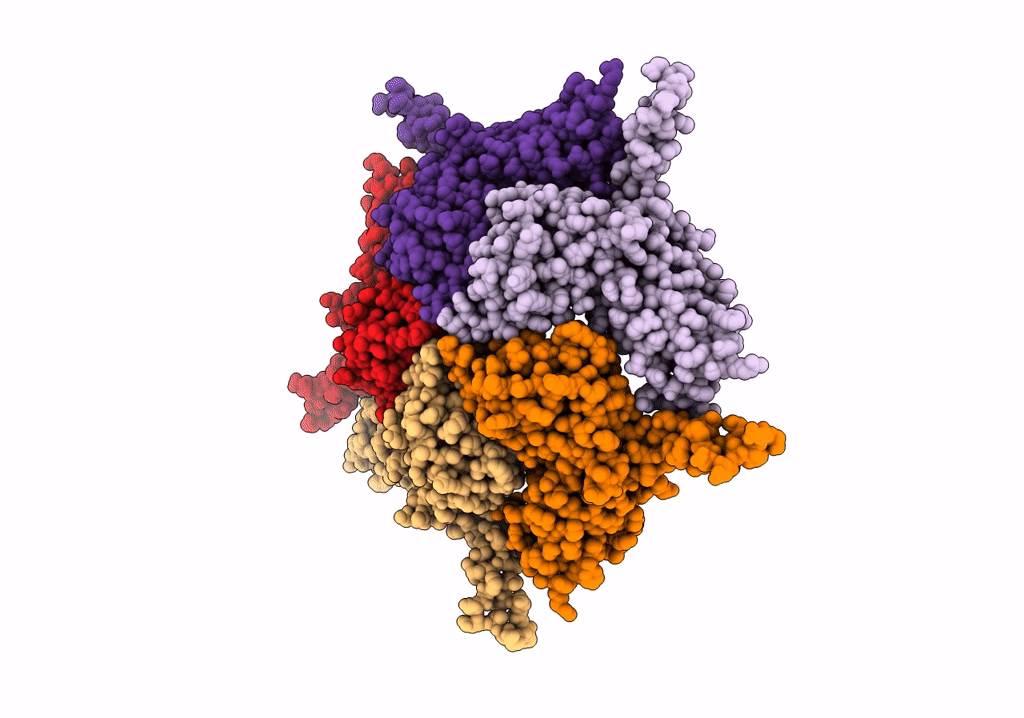 |
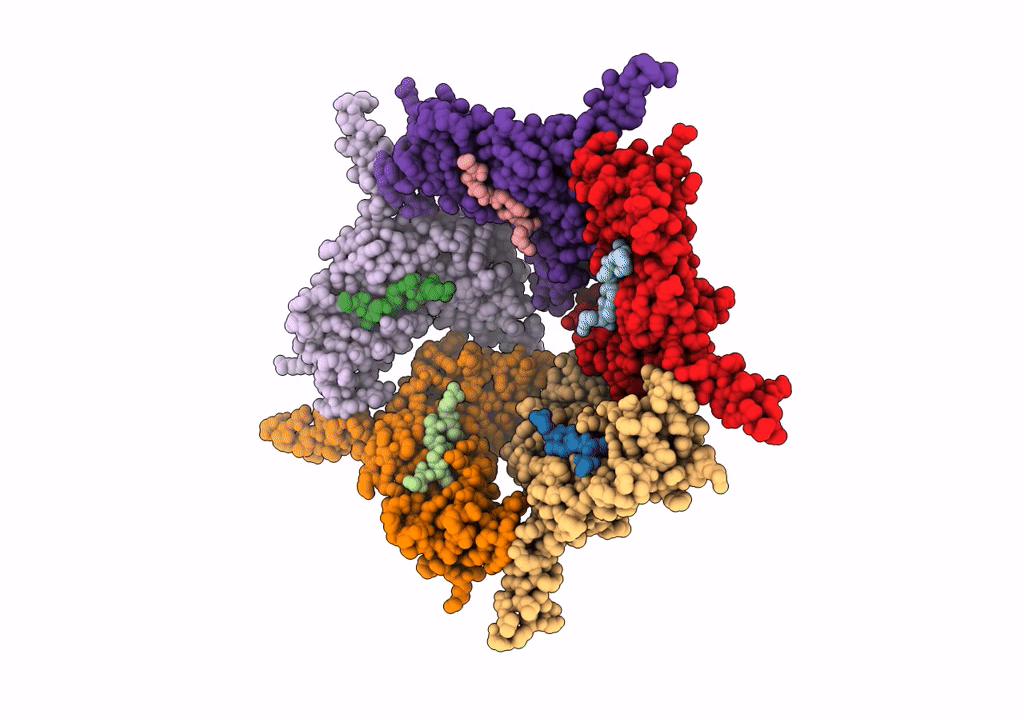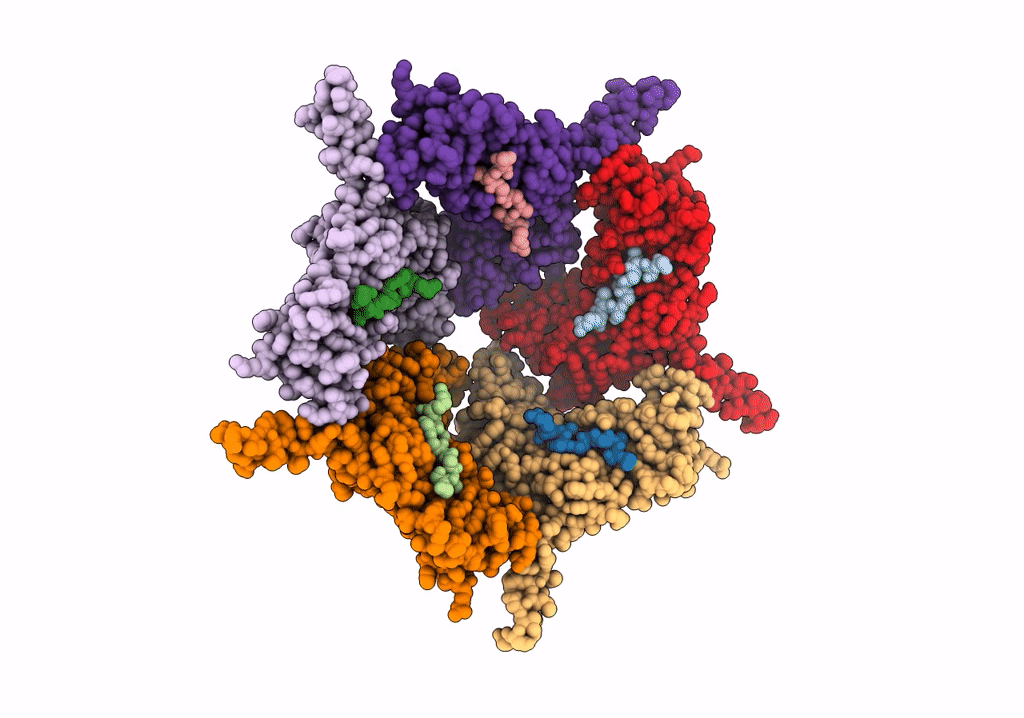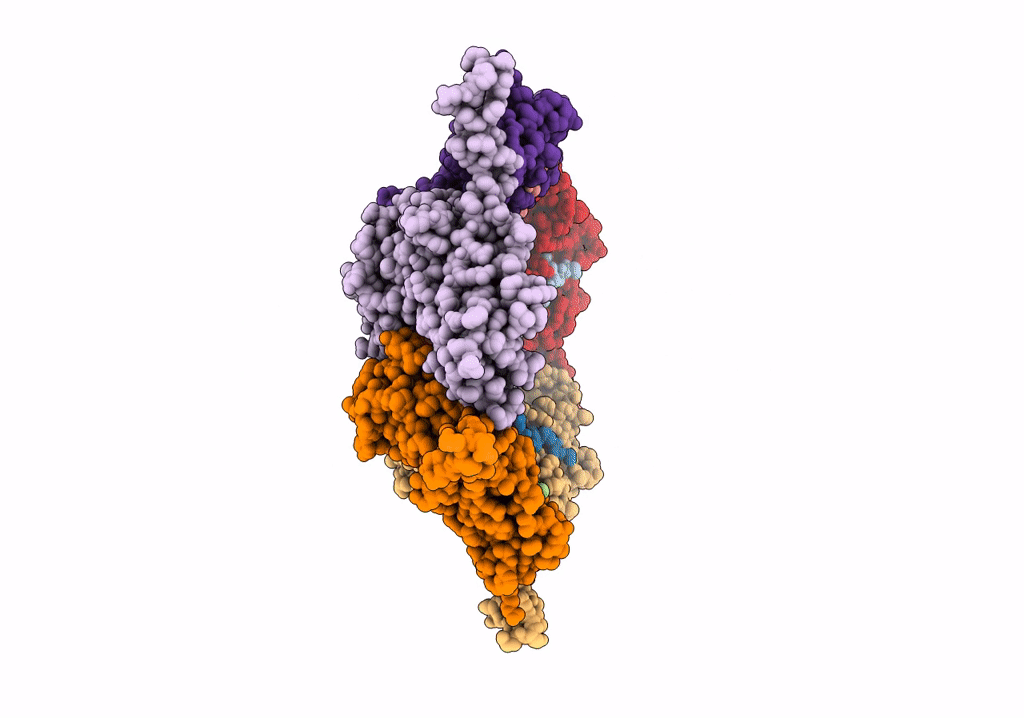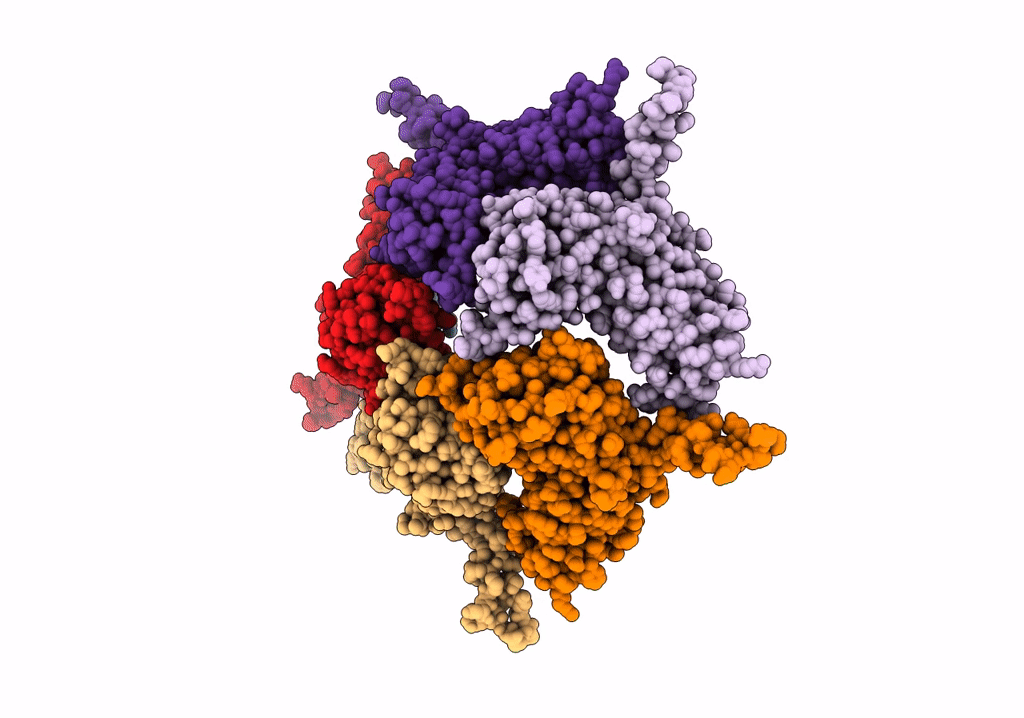
Model Of Closed Pentamer Of The Haliangium Ochraceum Encapsulin From Symmetry Expansion Of Icosahedral Single Particle Reconstruction
Organism: Haliangium ochraceum (strain dsm 14365 / jcm 11303 / smp-2)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: VIRUS LIKE PARTICLE |
 |
Model Of Open Pentamer Of The Haliangium Ochraceum Encapsulin From Symmetry Expansion Of Icosahedral Single Particle Reconstruction
Organism: Haliangium ochraceum (strain dsm 14365 / jcm 11303 / smp-2)
Method: ELECTRON MICROSCOPY Release Date: 2022-02-09 Classification: VIRUS LIKE PARTICLE |
 |
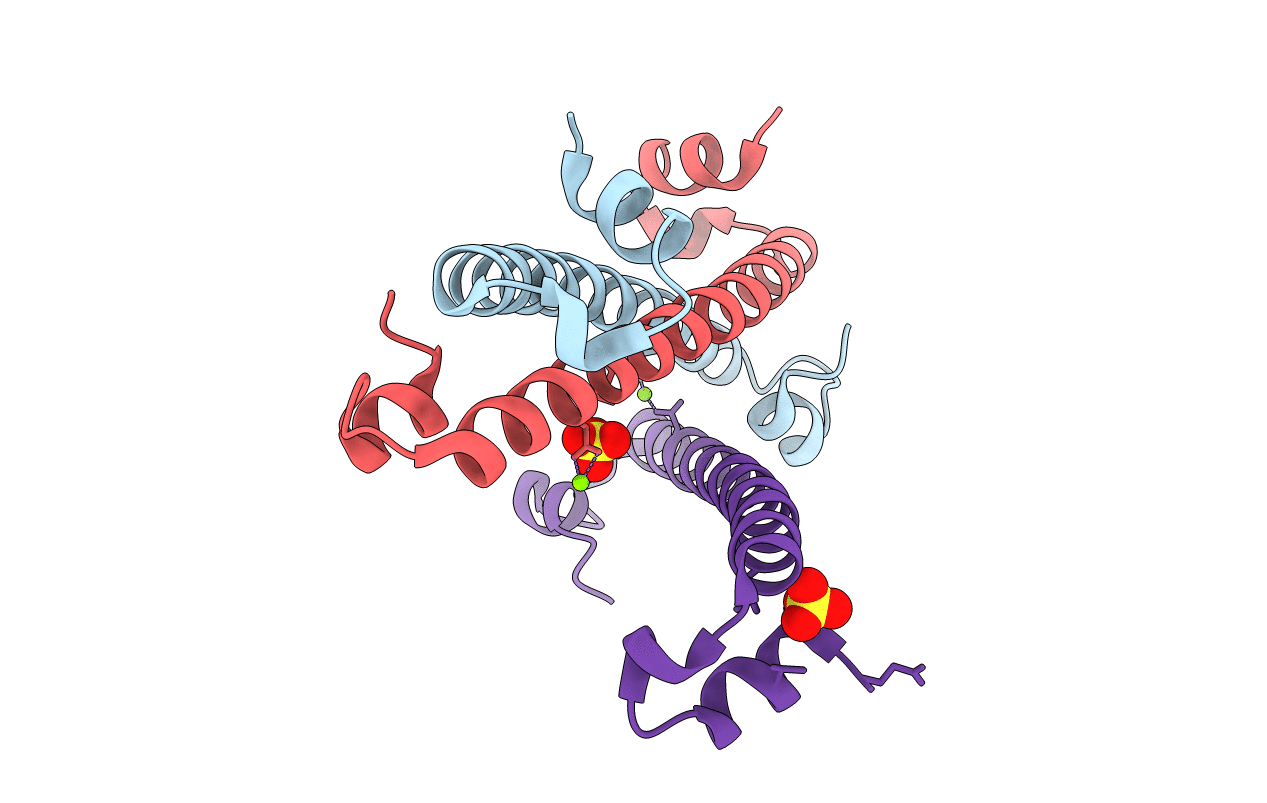
Organism: Rubrobacter radiotolerans
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-07-14 Classification: TRANSCRIPTION Ligands: MG, IPA |
 |
Organism: Rubrobacter radiotolerans
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2021-07-14 Classification: TRANSCRIPTION Ligands: SO4, MG |
 |
Organism: Rubrobacter radiotolerans
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-07-14 Classification: TRANSCRIPTION Ligands: C2E |
 |
Organism: Rhodospirillum rubrum (strain atcc 11170 / ath 1.1.1 / dsm 467 / lmg 4362 / ncib 8255 / s1)
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE Ligands: FE, CA |
 |
Organism: Rhodospirillum rubrum
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-09-23 Classification: OXIDOREDUCTASE Ligands: FE, CA |
 |
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745), Streptomyces sp. pansc19
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-11-13 Classification: TRANSCRIPTION Ligands: C2E |
 |
Structure Of S. Venezuelae Risg-Whig-C-Di-Gmp Complex: Orthorhombic Crystal Form
Organism: Streptomyces venezuelae (strain atcc 10712 / cbs 650.69 / dsm 40230 / jcm 4526 / nbrc 13096 / pd 04745), Streptomyces sp. pansc19
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-11-13 Classification: TRANSCRIPTION Ligands: C2E |
 |
Structure Of The Chromophore Binding Domain Of Stigmatella Aurantiaca Phytochrome P1, Wild-Type
Organism: Stigmatella aurantiaca dw4/3-1
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-09-19 Classification: SIGNALING PROTEIN Ligands: BLR |
 |
The Structure Of The Stigmatella Aurantiaca Phytochrome Chromophore Binding Domain T289H Mutant
Organism: Stigmatella aurantiaca dw4/3-1
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2018-09-19 Classification: SIGNALING PROTEIN Ligands: BLR |
 |
Organism: Stigmatella aurantiaca dw4/3-1
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2018-09-19 Classification: SIGNALING PROTEIN Ligands: BLR |

