Search Count: 20
 |
Organism: Pyrococcus horikoshii ot3
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-09-02 Classification: TRANSFERASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-08-29 Classification: SIGNALING PROTEIN/TRANSFERASE Ligands: ZN, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2017-09-06 Classification: SIGNALING PROTEIN Ligands: MG, ARG |
 |
Organism: Lactobacillus brevis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2017-08-02 Classification: LYASE Ligands: PLP |
 |
Organism: Palmaria palmata
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2016-10-05 Classification: PHOTOSYNTHESIS Ligands: CYC, PUB |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.81 Å Release Date: 2016-04-20 Classification: HYDROLASE INHIBITOR/PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-03-09 Classification: CELL ADHESION/PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-03-09 Classification: CELL ADHESION/PROTEIN BINDING Ligands: EDO |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-03-09 Classification: PROTEIN BINDING/CELL ADHESION |
 |
Organism: Todarodes pacificus
Method: X-RAY DIFFRACTION Release Date: 2015-10-14 Classification: OXYGEN TRANSPORT Ligands: CUO |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, PRU |
 |
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant D202N In Complex With Glucose And Glycerol
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL, BGC |
 |
Crystal Structure Of Alpha-Glucosidase Mutant E271Q In Complex With Maltose
Organism: Halomonas sp. h11
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2015-06-10 Classification: HYDROLASE Ligands: MG, GOL |
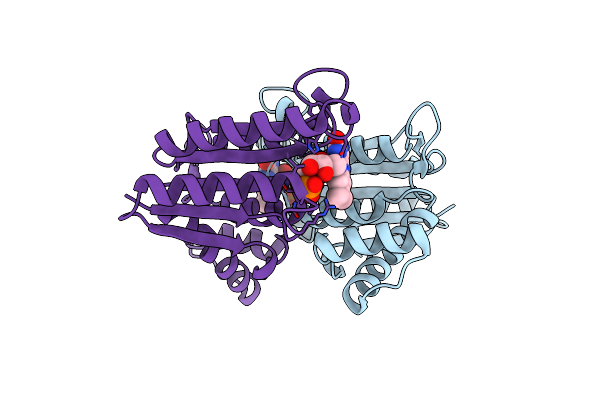 |
Organism: Bacillus
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2014-02-12 Classification: OXIDOREDUCTASE Ligands: FMN |
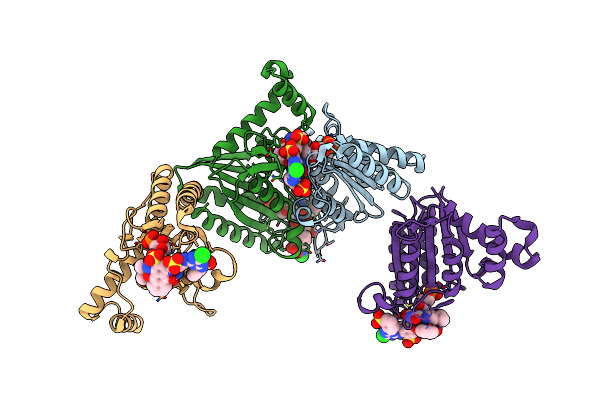 |
Crystal Structure Of Azoreductase Azrc In Complex With Nad(P)-Inhibitor Cibacron Blue
Organism: Bacillus
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2014-02-12 Classification: OXIDOREDUCTASE/ OXIDOREDUCTASE INHIBITOR Ligands: FMN, CBD |
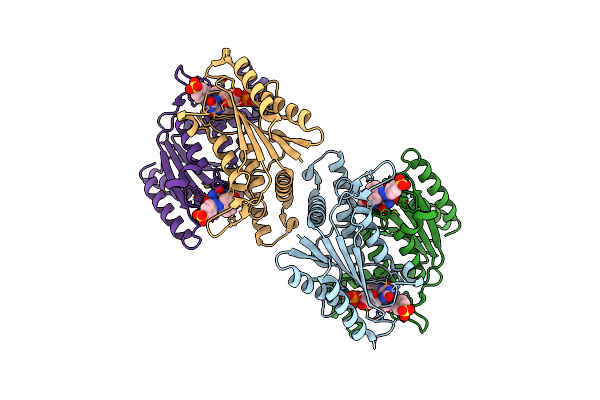 |
Crystal Structure Of Azoreductase Azrc In Complex With Sulfone-Modified Azo Dye Orange I
Organism: Bacillus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2014-02-12 Classification: OXIDOREDUCTASE Ligands: FMN, ORI |
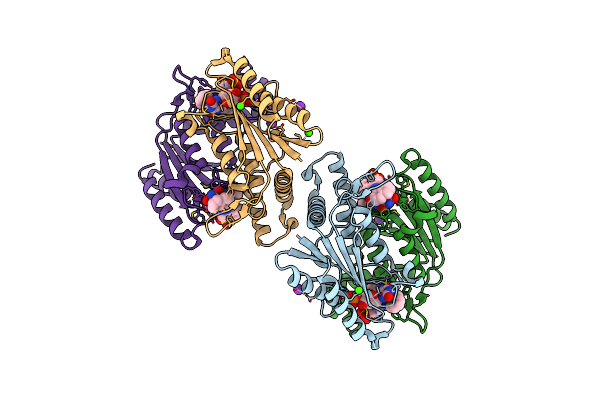 |
Crystal Structure Of Azoreductase Azrc Fin Complex With Sulfone-Modified Azo Dye Acid Red 88
Organism: Bacillus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-02-12 Classification: OXIDOREDUCTASE Ligands: FMN, RE8, CA, K |
 |
Organism: Unidentified prokaryotic organism
Method: X-RAY DIFFRACTION Resolution:1.03 Å Release Date: 2013-10-30 Classification: TRANSPORT PROTEIN Ligands: PO4, GOL |
 |
Organism: Unidentified prokaryotic organism
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2013-10-30 Classification: TRANSPORT PROTEIN Ligands: PO4, GOL |

