Search Count: 37
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2024-06-12 Classification: FLAVOPROTEIN Ligands: PO4 |
 |
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN |
 |
Organism: Lactiplantibacillus plantarum
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN Ligands: MN, PO4 |
 |
Organism: Lactobacillus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: FLAVOPROTEIN |
 |
Crystal Structure Of Phenazine 1-Carboxylic Acid Decarboxylase From Mycobacterium Fortuitum
Organism: Mycolicibacterium fortuitum, Unidentified
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2022-08-24 Classification: LYASE Ligands: MN, 4LU, NA |
 |
Crystal Structure Of Indole 3-Carboxylic Acid Decarboxylase From Arthrobacter Nicotianae Fi1612 In Complex With Co-Factor Prfmn.
Organism: Glutamicibacter nicotianae
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2022-03-02 Classification: LYASE |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.07 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, FIV, PEG |
 |
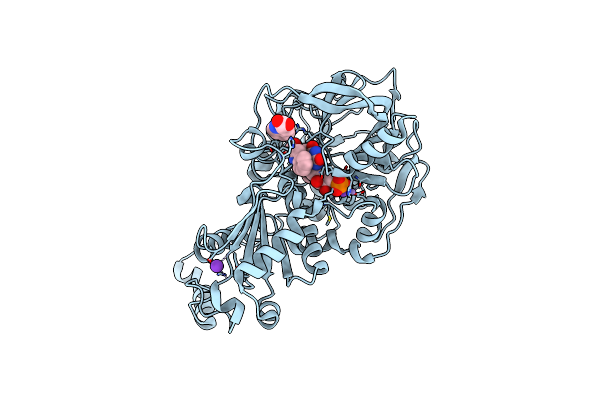
Structure Of A. Niger Fdc I327S Variant In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
 |
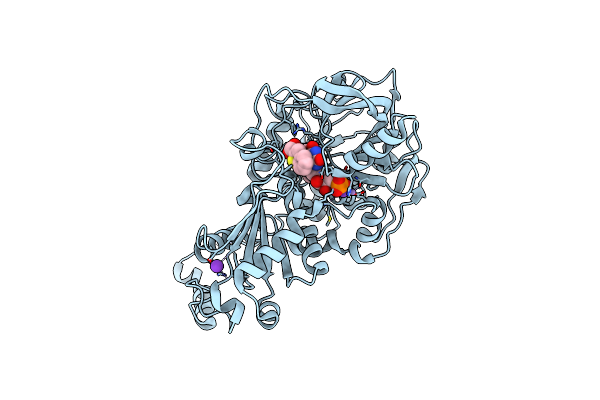
Structure Of A. Niger Fdc I327S Variant In Complex With Indol-2-Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.06 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
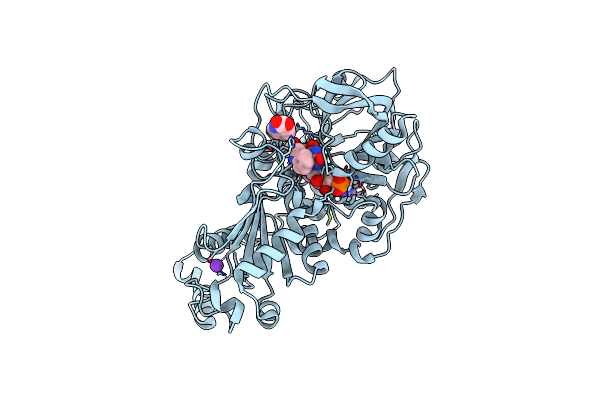
Structure Of A. Niger Fdc Wt In Complex With Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.02 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ND8 |
 |
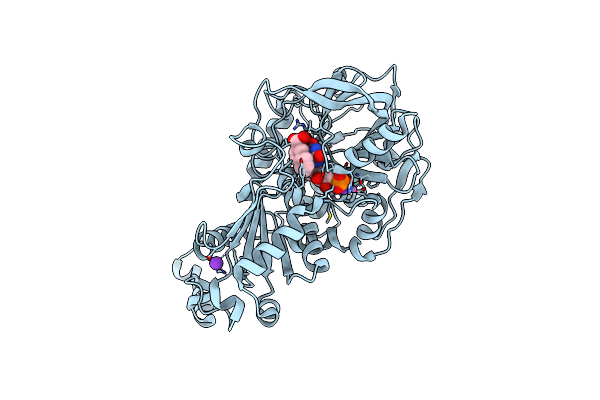
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.12 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, BYN, ICB |
 |
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, FIV |
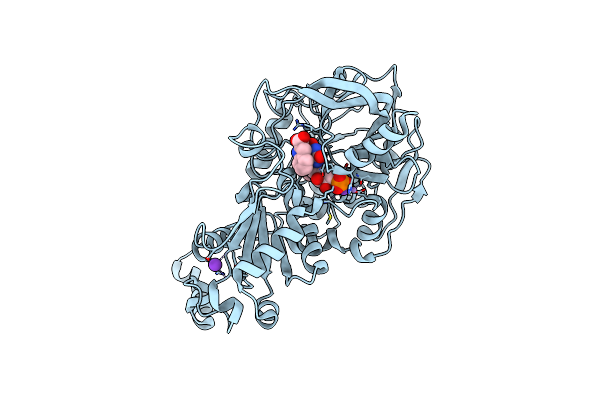 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Indole 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ICB |
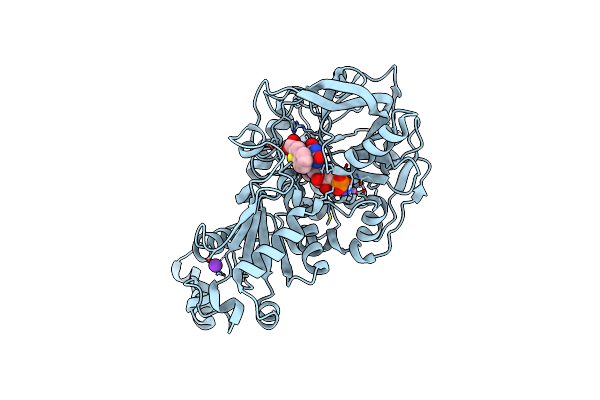 |
Structure Of A. Niger Fdc Wt In Complex With Fmn And Benzothiophene 2 Carboxylic Acid
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-06-24 Classification: LIGASE Ligands: MN, K, FMN, ND8 |
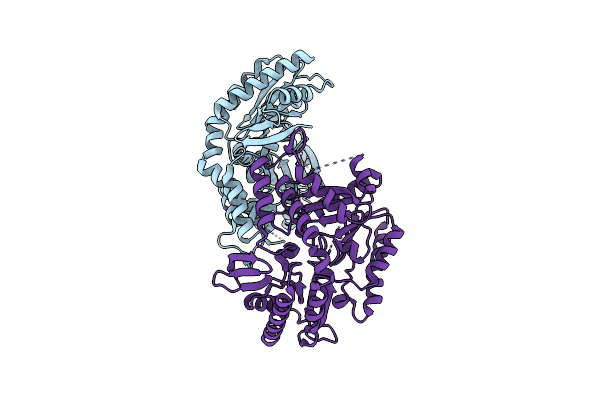 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-06-12 Classification: TRANSFERASE |
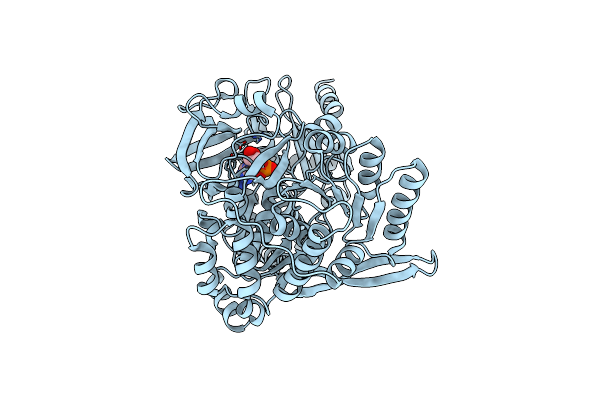 |
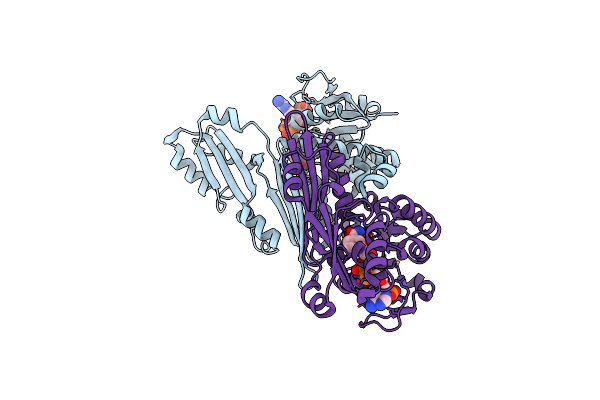
Structure Of The A Domain Of Carboxylic Acid Reductase (Car) From Nocardia Iowensis In Complex With Amp And Iodide
Organism: Nocardia iowensis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2018-01-17 Classification: OXIDOREDUCTASE Ligands: AMP, IOD |
 |
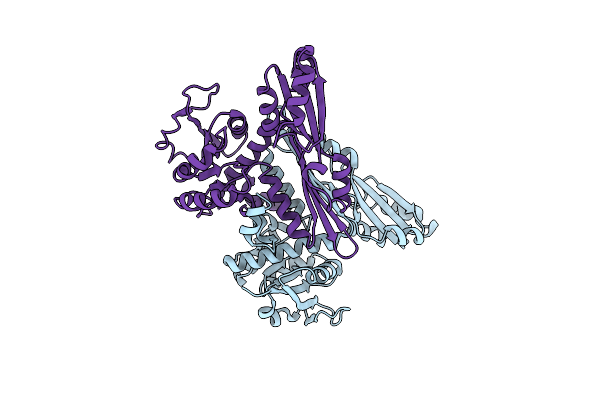
Crystal Structure Of The Engineered D-Amino Acid Dehydrogenase (Daadh) Bound To Nadp+
Organism: Corynebacterium glutamicum (strain atcc 13032 / dsm 20300 / jcm 1318 / lmg 3730 / ncimb 10025)
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
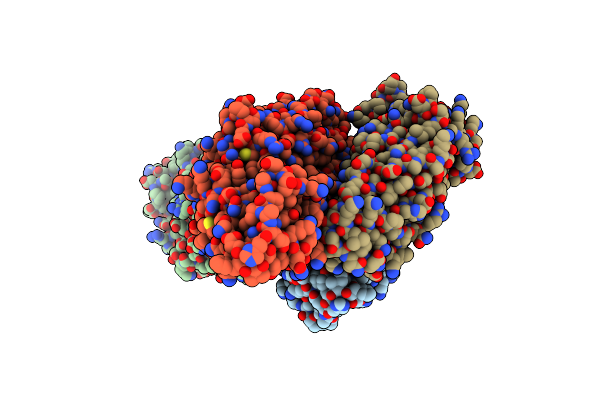
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2017-08-16 Classification: OXIDOREDUCTASE |
 |
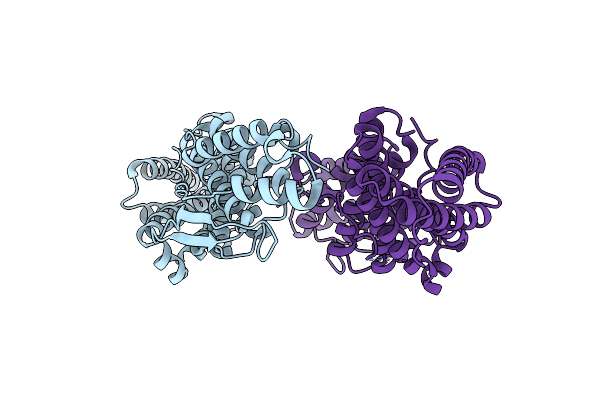
Crystal Structure Of The His Domain Protein Tyrosine Phosphatase (Hd-Ptp/Ptpn23) Bro1 Domain (Sara Complex Structure)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-08-16 Classification: HYDROLASE |
 |
Crystal Structure Of The His Domain Protein Tyrosine Phosphatase (Hd-Ptp/Ptpn23) Bro1 Domain (Apo Structure)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2017-08-16 Classification: HYDROLASE |

