Search Count: 16
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture -Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol 30:20:20:30)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/-Sterol (Dopc, Dope, Dops, Pi(4,5)P2 50:20:20:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P |
 |
Helical Reconstruction Of Yeast Eisosome Protein Pil1 Bound To Membrane Composed Of Lipid Mixture +Pip2/+Sterol (Dopc, Dope, Dops, Cholesterol, Pi(4,5)P2 35:20:20:15:10)
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P, P5S |
 |
Compact State - Pil1 In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Compact State - Pil1 Dimer With Lipid Headgroups Fitted In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN Ligands: I3P, SEP |
 |
Stretched State - Pil1 In Native Eisosome Lattice Bound To Plasma Membrane Microdomain
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-07-24 Classification: LIPID BINDING PROTEIN |
 |
Cryo-Em Structure Of Saccharomyces Cerevisiae Toroid (Torc1 Organized In Inhibited Domains).
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2023-01-18 Classification: SIGNALING PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN Ligands: ZN |
 |
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2022-11-02 Classification: SIGNALING PROTEIN |
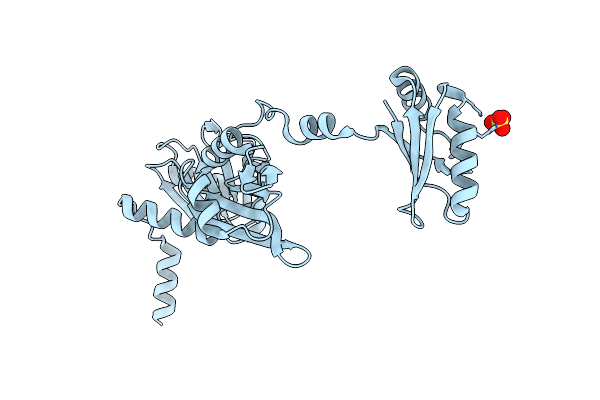 |
The Crystal Structure Of The Split End Protein Sharp Adds A New Layer Of Complexity To Proteins Containing Rna Recognition Motifs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-05-14 Classification: TRANSCRIPTION Ligands: SO4 |
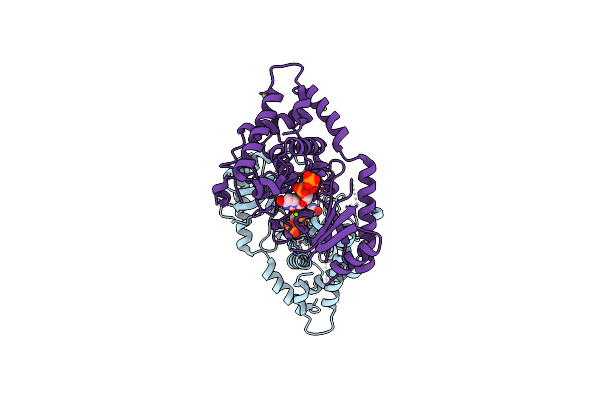 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-18 Classification: TRANSFERASE Ligands: 2KH, BR, MG |
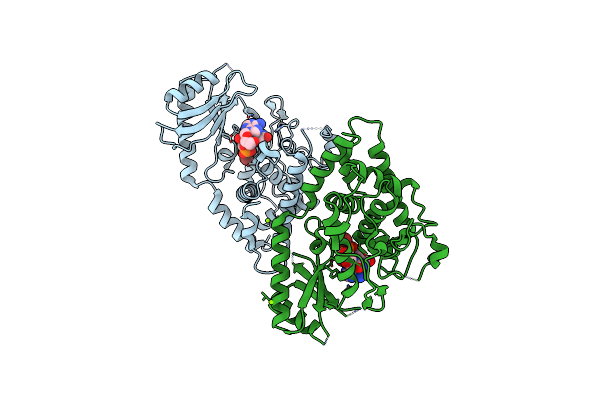 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2013-12-18 Classification: TRANSFERASE/RNA Ligands: MG, BR |
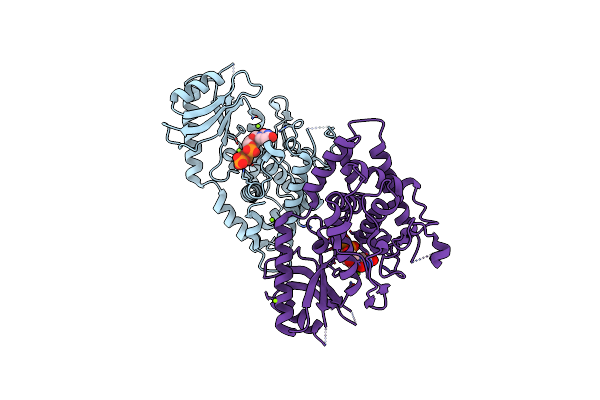 |
Organism: Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:2.28 Å Release Date: 2012-06-06 Classification: TRANSFERASE Ligands: UTP, MG |
 |
Structural Basis Of Transcriptional Gene Silencing Mediated By Arabidopsis Mom1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2012-03-07 Classification: TRANSCRIPTION |

