Search Count: 2,766
All
Selected
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: C5P, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: C5P, KDO, GLY, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2026-01-28 Classification: TRANSFERASE Ligands: C5P, KDO, CL |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Substrate-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Resolution:2.54 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Catalysis And Product-Bound State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG, UDP, A1B94 |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Apo State (Octamer Volume)
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Resolution:2.79 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN |
 |
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Cryo-Em Structure Of The Glycosyltransferase Gtrb In The Pre-Intermediate State
Organism: Synechocystis sp. pcc 6803 substr. kazusa
Method: ELECTRON MICROSCOPY Resolution:2.98 Å Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN Ligands: UPG, 5TR, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2025-12-10 Classification: SUGAR BINDING PROTEIN Ligands: MN, MLI |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: Y01, MN, LMT |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, MN, Y01 |
 |
Organism: Paramecium bursaria chlorella virus cz-2, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: TRANSFERASE/IMMUNE SYSTEM Ligands: UGA, Y01, MN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: TRANSFERASE Ligands: NAG, UDP, MN |
 |
Organism: Trifolium subterraneum
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Trifolium subterraneum
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: A1L68, BMA, UDP |
 |
Udp-Binding Psbglut, A Tetrahydrobiopterin Glucosyltransferase From Pseudanabaena Sp. Chao 1811
Organism: Pseudanabaena sp. chao 1811
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: UDP |
 |
Organism: Schisandra chinensis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-11-19 Classification: PLANT PROTEIN Ligands: UDP, GOL |
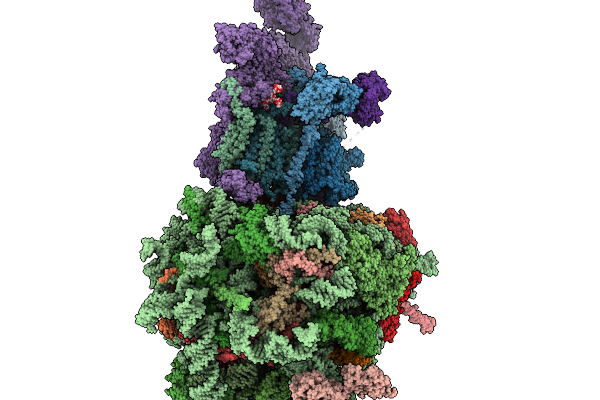 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU, MG, ZN |
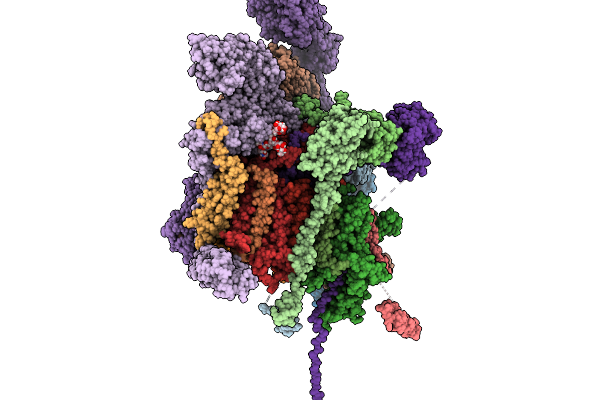 |
Structure Of A Grp94 Folding Intermediate Engaged With A Ccdc134- And Fkbp11-Bound Secretory Translocon
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: RIBOSOME Ligands: ELU |
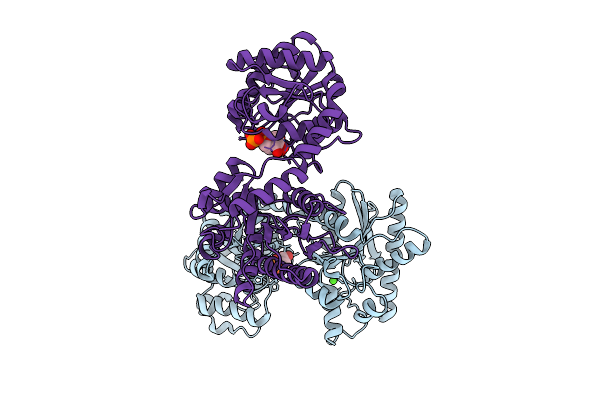 |
Organism: Leishmania major
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2025-10-29 Classification: SUGAR BINDING PROTEIN Ligands: CA, UDP |

