Search Count: 101
All
Selected
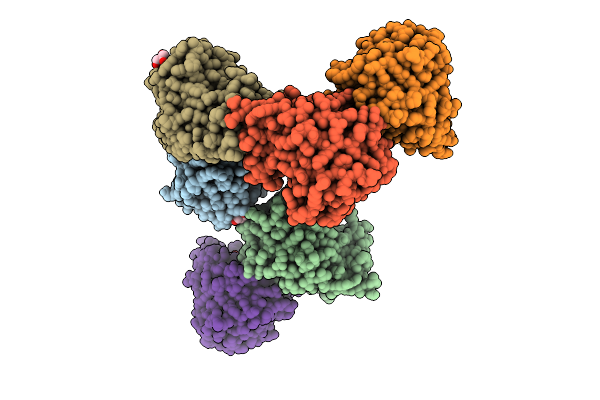 |
Crystal Structure Of Bacteroides Ovatus Kdui1 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-10-29 Classification: ISOMERASE Ligands: GOL |
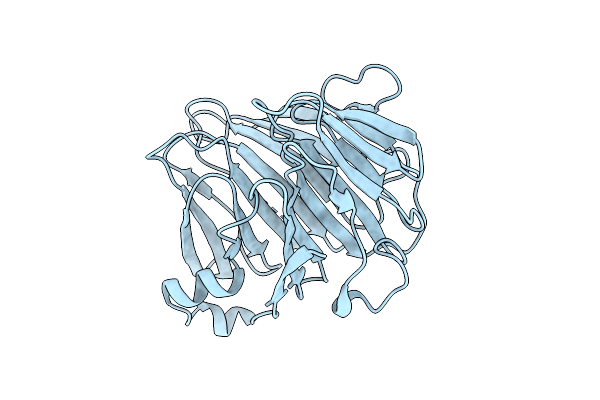 |
Crystal Structure Of Bacteroides Ovatus Kdui2 Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2025-10-29 Classification: ISOMERASE |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
Crystal Structure Of Bacteroides Ovatus Dhud Complexed With Nad+ Responsible For Metabolism Of Glycosaminoglycan
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-10-29 Classification: OXIDOREDUCTASE Ligands: NAD |
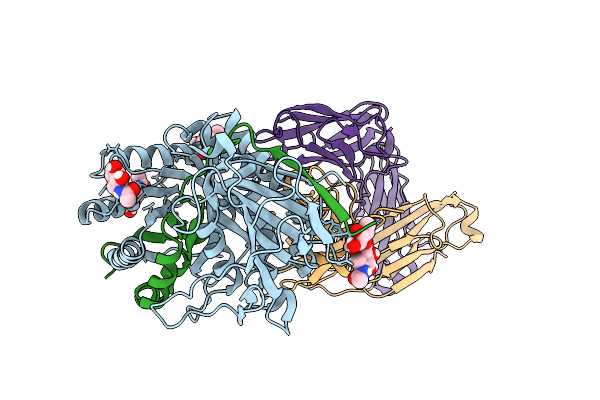 |
Organism: Homo sapiens, Mus musculus
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2025-09-24 Classification: CARBOHYDRATE Ligands: NAG |
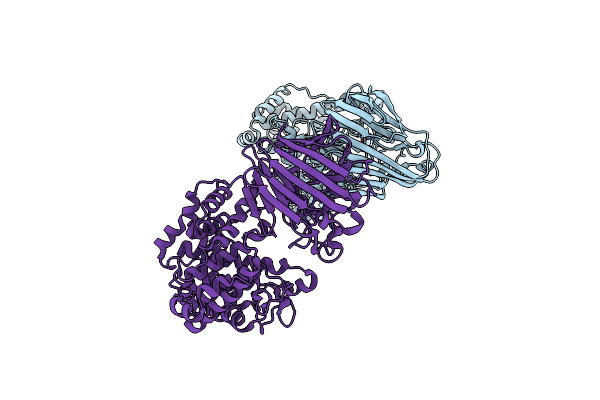 |
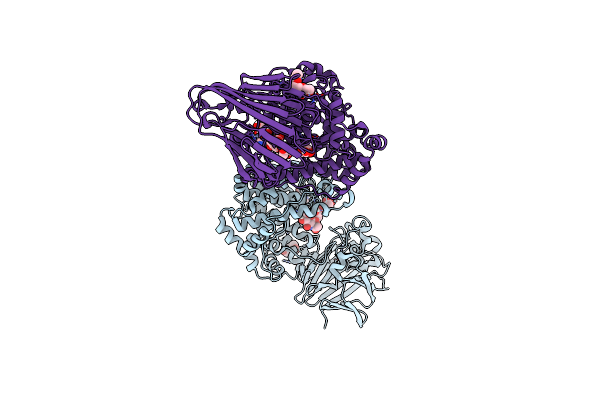
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
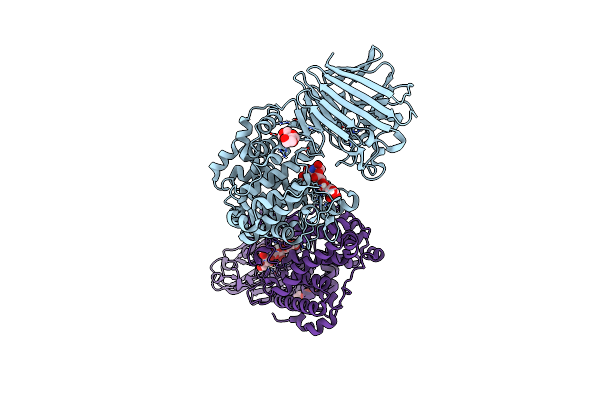
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-12-04 Classification: LYASE Ligands: P4K, MES, ZN |
 |
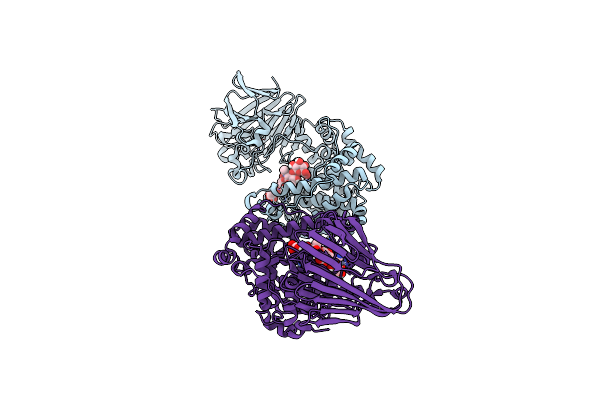
Polysaccharide Lyase Btpl33Ha (Bt4410) Y291A With Hadp4 Collected At 1.22 A
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2024-12-04 Classification: LYASE Ligands: P4K, ZN |
 |
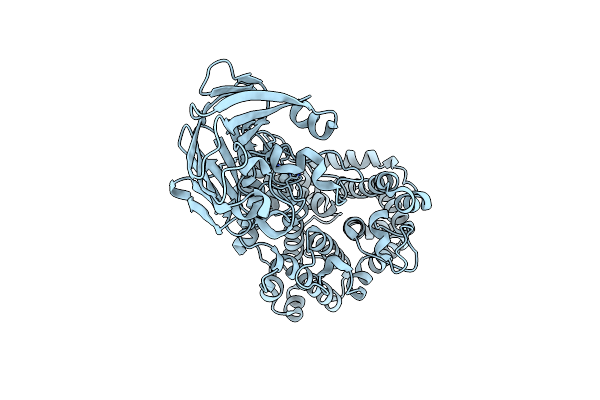
Polysaccharide Lyase Btpl33Ha (Bt4410) Y291A With Ha Dp4 Collected At 1.33 A
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Organism: Bacteroides cellulosilyticus wh2
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2024-12-04 Classification: LYASE Ligands: ZN |
 |
Organism: Spirosoma fluviale
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-09-04 Classification: LYASE Ligands: MN, CA |
 |
Organism: Bacteroides intestinalis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-09-04 Classification: LYASE Ligands: MN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-07-31 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-02-28 Classification: SIGNALING PROTEIN Ligands: SO4, MAN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-02-28 Classification: SIGNALING PROTEIN Ligands: MAN, NO3 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.56 Å Release Date: 2024-02-28 Classification: SIGNALING PROTEIN Ligands: MAN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2024-02-28 Classification: SIGNALING PROTEIN |
 |
Human Heparan Sulfate N-Deacetylase-N-Sulfotransferase 1 In Complex With Calcium, 3'-Phosphoadenosine-5'-Phosphosulfate And Nanobody Nab13 (Composite Map And Model).
Organism: Lama glama, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-21 Classification: CARBOHYDRATE Ligands: A3P, CA |
 |
Human Heparan Sulfate N-Deacetylase-N-Sulfotransferase 1 In Complex With Calcium And 3'-Phosphoadenosine-5'-Phosphosulfate
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-02-07 Classification: CARBOHYDRATE Ligands: A3P, CA |

