Search Count: 277
 |
Glyceraldehyde 3-Phosphate Dehydrogenase A (Gapdha) Apoenzyme, From Helicobacter Pylori
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SO4 |
 |
Glyceraldehyde 3-Phosphate Dehydrogenase A (Gapdha) Nadp Holoenzyme, From Helicobacter Pylori, With Active Site Cysteine Oxidised To Sulfenic Acid
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
 |
Crystal Structure Of Bacillus Subtilis Glyceraldehyde-3-Phosphate Dehydrogenase Gapb
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-02 Classification: OXIDOREDUCTASE |
 |
Structure Of The Class Ii Fructose-1,6-Bisphophatase From Francisella Tularensis Complexed With Native Metal Cofactor Mn++ And Product F6P
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2023-06-28 Classification: HYDROLASE/PRODUCT Ligands: MN, F6P |
 |
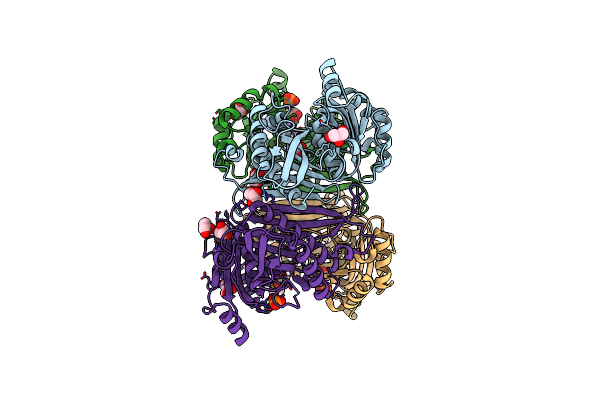
Structure Of The Class Ii Fructose-1,6-Bisphophatase From Mycobacterium Tuberculosis Complexed With Substrate F1,6Bp
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:3.71 Å Release Date: 2023-06-28 Classification: HYDROLASE/SUBSTRATE Ligands: GOL, FBP, MG |
 |
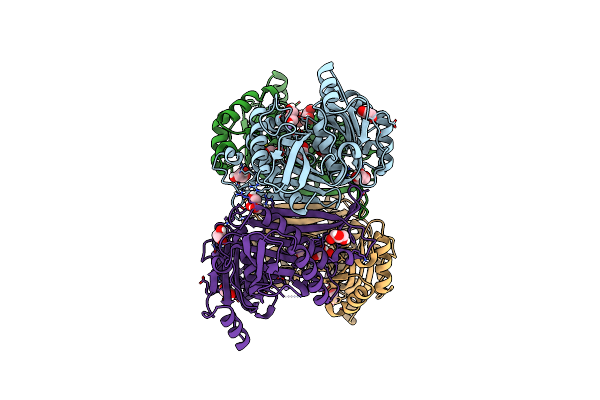
Structure Of The Class Ii Fructose-1,6-Bisphosphatase From Francisella Tularensis With Native Mn++ Divalent Cation And Partially Occupied Product F6P
Organism: Francisella tularensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-06-28 Classification: HYDROLASE Ligands: GOL, MN, PO4 |
 |
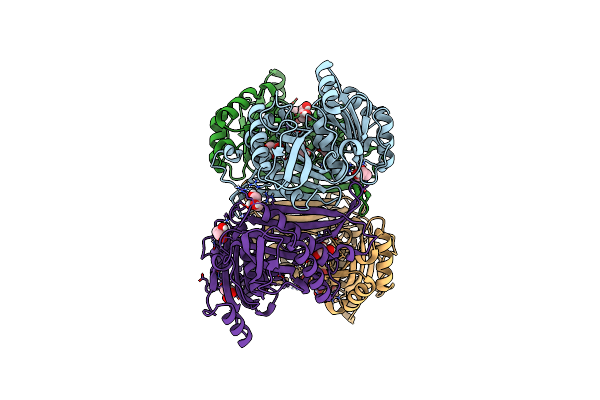
Structure Of The Class Ii Fructose-1,6-Bisphophatase From Francisella Tularensis Complexed With Native Metal Cofactor Mn++
Organism: Francisella cf. tularensis subsp. novicida 3523
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-06-28 Classification: HYDROLASE Ligands: GOL, MN |
 |
Structure Of The Class Ii Fructose-1,6-Bisphophatase From Francisella Tularensis Complexed With Native Metal Cofactor Mn++ And Substrate Fructose-1,6-Bisphosphate
Organism: Francisella cf. tularensis subsp. novicida 3523
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-06-28 Classification: HYDROLASE Ligands: GOL, MN, FBP |
 |
Crystal Structure Of Cytoplasmic Triosephosphate Isomerase From Cuscuta Australis
Organism: Cuscuta australis
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2023-05-17 Classification: ISOMERASE Ligands: CL |
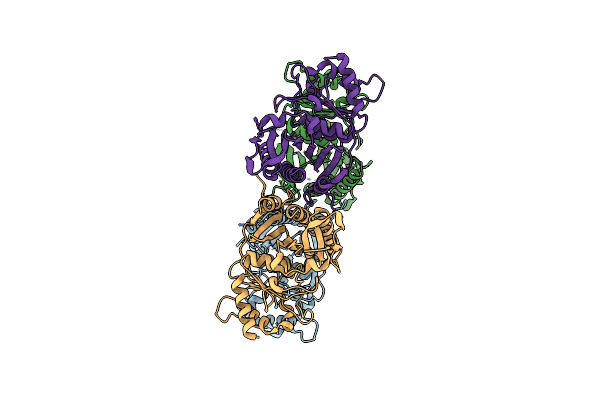 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2023-02-15 Classification: HYDROLASE |
 |
Crystal Structure Of Chloroplast Triosephosphate Isomerase From Cuscuta Australis
Organism: Cuscuta australis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-10-26 Classification: ISOMERASE Ligands: ACT |
 |
Crystal Structure Of Triosephosphate Isomerase From Verrucomicrobium Spinosum
Organism: Verrucomicrobium spinosum
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2022-08-10 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase From Bacteroides Thetaiotaomicron
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-08-10 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidatus Absconditabacteria (Sr1) Bacterium
Organism: Candidate division sr1 bacterium raac1_sr1_1
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-07-20 Classification: ISOMERASE |
 |
Crystal Structure Of Triosephosphate Isomerase From Ktedonobacter Racemifer
Organism: Ktedonobacter racemifer
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-07-13 Classification: ISOMERASE Ligands: NO3 |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidate Division Katanobacteria (Wwe3) Bacterium
Organism: Candidate division wwe3 bacterium raac2_wwe3_1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-07-06 Classification: ISOMERASE Ligands: PO4 |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidatus Roizmanbacteria
Organism: Candidatus roizmanbacteria
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-07-06 Classification: ISOMERASE Ligands: PO4 |
 |
Crystal Structure Of Triosephosphate Isomerase From Candidatus Prometheoarchaeum Syntrophicum
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2022-06-15 Classification: ISOMERASE |
 |
Glyceraldehyde 3-Phosphate Dehydrogenase From Campylobacter Jejeuni - Nad(P) Complex
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: NAP, PEG, NA |
 |
Glyceraldehyde 3-Phosphate Dehydrogenase From Campylobacter Jejeuni - Adp Complex
Organism: Campylobacter jejuni subsp. jejuni serotype o:2 (strain atcc 700819 / nctc 11168)
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2022-01-12 Classification: OXIDOREDUCTASE Ligands: SO4, ADP, BME, ACT |

