Search Count: 1,883
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: DNA BINDING PROTEIN Ligands: ACY, SO4, HCI |
 |
Organism: Caenorhabditis elegans
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: GENE REGULATION Ligands: SO4 |
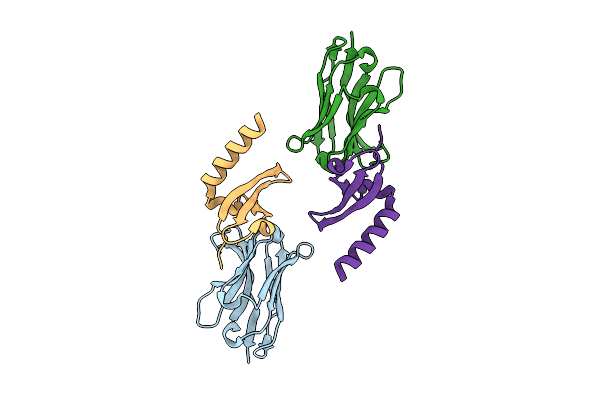 |
Organism: Lama glama, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION |
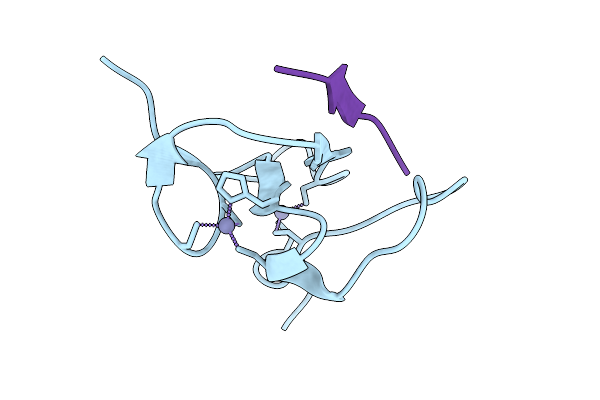 |
Crystal Structure Of Arabidopsis Thaliana Ing1 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
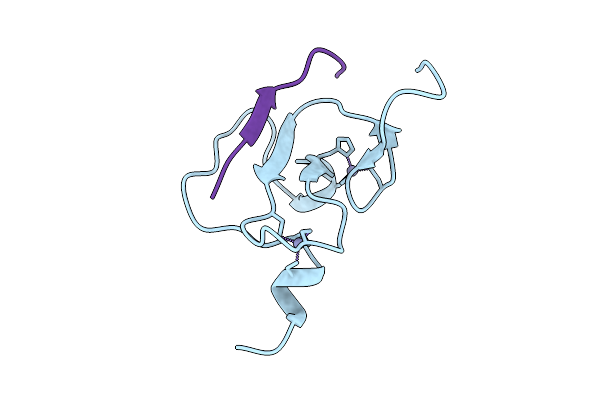 |
Crystal Structure Of Arabidopsis Thaliana Ing2 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
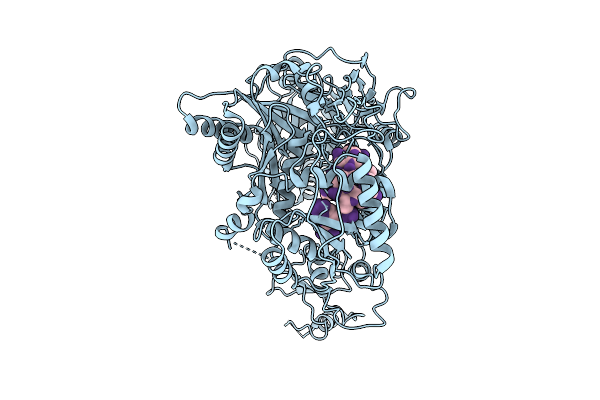 |
Structure Of Neurodevelopmental Mutant Ago1 F180Del In Complex With Guide Rna
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: GENE REGULATION |
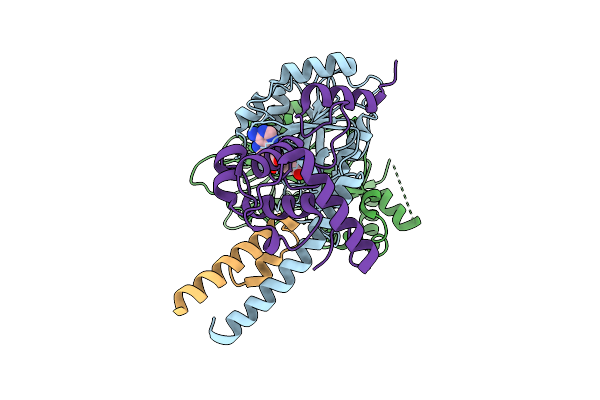 |
Organism: Tetrahymena thermophila sb210
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN Ligands: SAM |
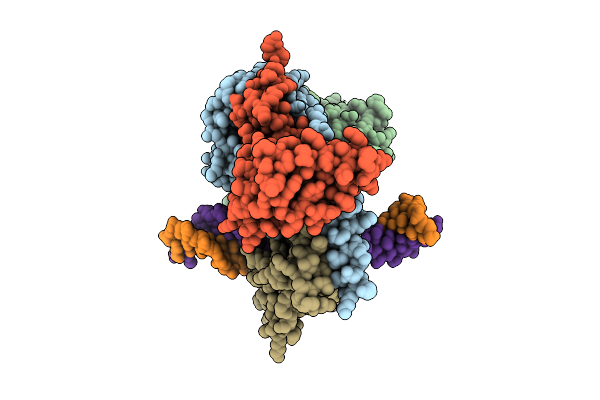 |
Cryo-Em Structure Of Hemimethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
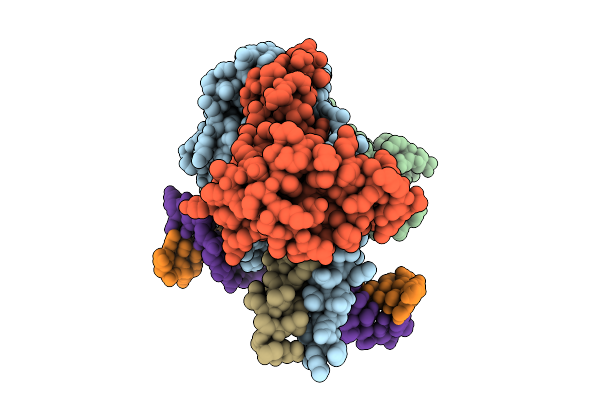 |
Cryo-Em Structure Of Hemi-Methylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
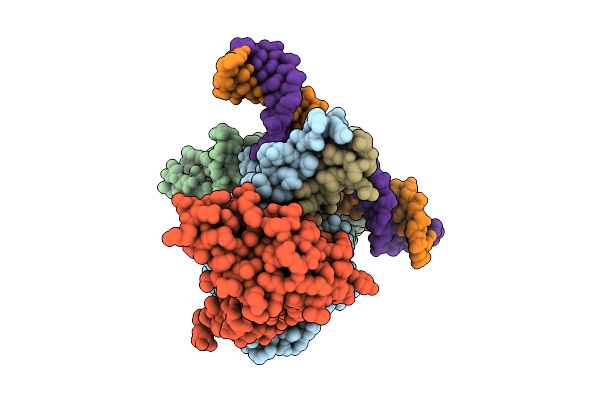 |
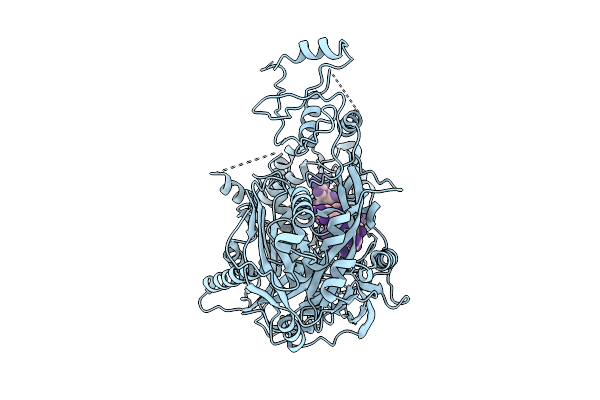
Cryo-Em Structure Of Unmethylated Dna-Bound Tetrahymena Dna Methyltransferase Complex Mta1C (Mta9-B)
Organism: Tetrahymena thermophila sb210, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-11-05 Classification: DNA BINDING PROTEIN/DNA Ligands: SAM |
 |
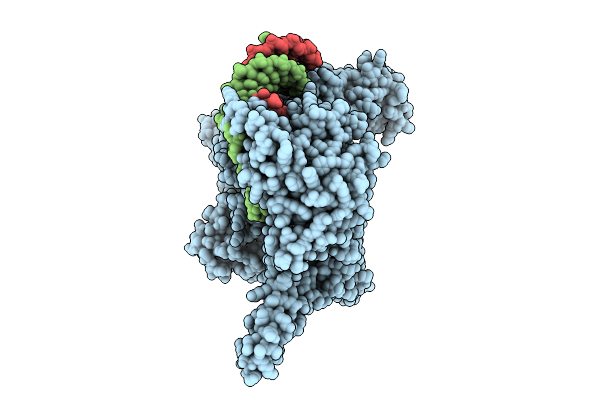
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: GENE REGULATION |
 |
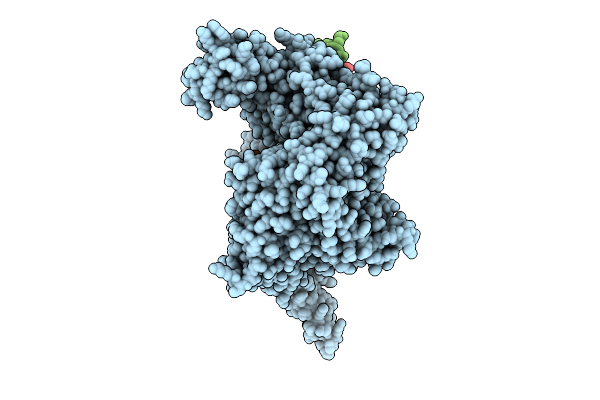
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
 |
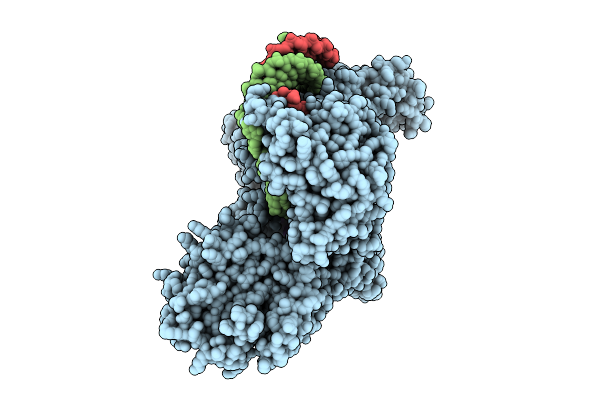
Structure Of Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid Bound To Dntp And Mn2+
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: DTP, ZN, MN |
 |
Structure Of Full-Length Human Line-1 Orf2P With Endogenous Dna And Rna/Cdna Hybrid
Organism: Homo sapiens, Trichoplusia ni
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: GENE REGULATION Ligands: ZN |
 |
E3 Ubiquitin Ligase Huwe1 Homolog Tom1P In Closed-Conformation With Internal Acidic Domain Deletion
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: GENE REGULATION |
 |
Full-Length And Internally His-Tagged Yeast E3 Ubiquitin Ligase Tom1P In An Open-Conformation
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: TRANSFERASE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-01 Classification: GENE REGULATION |
 |
Cryo-Em Structure Of Full-Length And Internally His-Tagged Yeast E3 Ubiquitin Ligase Tom1P, In A Closed-Conformation State With Helical Repeat Solenoid Architecture
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: GENE REGULATION |
 |
Crystal Structure Of The Lsd1/Corest Histone Demethylase In Complex With The Cofactor Fad And The Inhibitor Gsk690
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-09-17 Classification: GENE REGULATION Ligands: FAD, GOL, A1A5U |
 |
Organism: Cricetulus griseus
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: NUCLEAR PROTEIN |

