Search Count: 175
 |
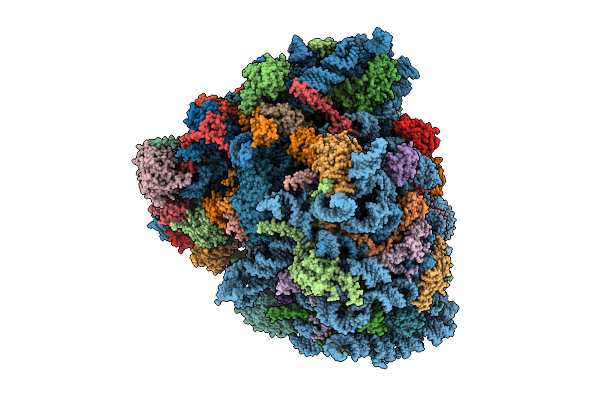
Gtpbp1*Gcp*Phe-Trna*Ribosome In The Gtpase Activation-Like State, Structure Iii
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
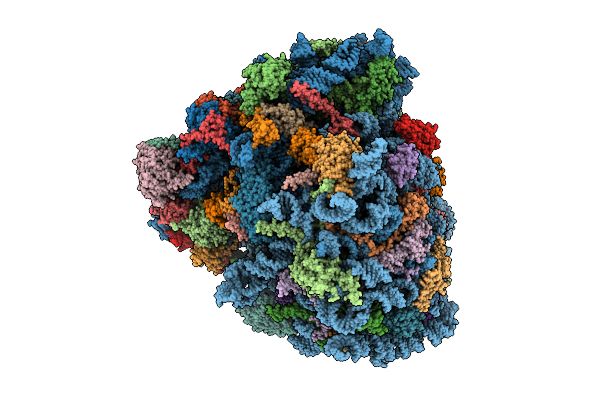
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
 |
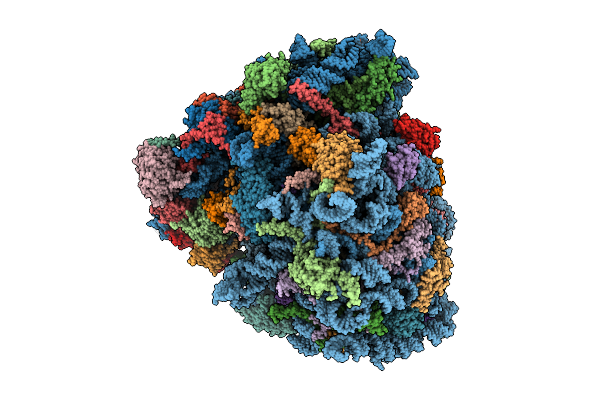
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
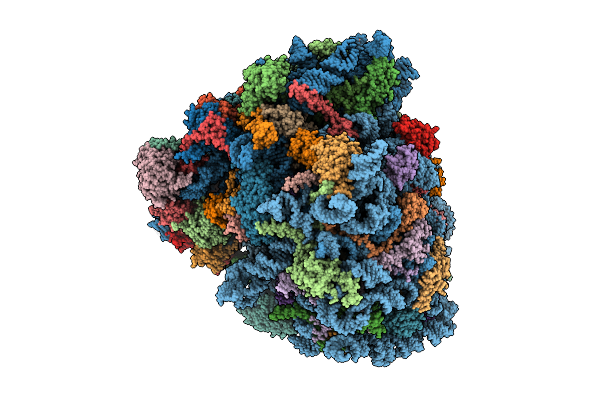
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, GCP, MG, K |
 |
Organism: Homo sapiens, Oryctolagus cuniculus, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: RIBOSOME Ligands: SPD, ZN, GTP, PHE, ATP, MET, K, GCP, MG |
 |
Crystal Structure Of Compound 1-Mediated Ternary Complex Of Kras G12R Gcp With Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2025-11-12 Classification: ONCOPROTEIN Ligands: GOL, X53, GCP, MG, FLC |
 |
Crystal Structure Of Compound 3-Mediated Ternary Complex Of Kras G12R Gcp With Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2025-11-12 Classification: ONCOPROTEIN Ligands: FLC, A1JH1, GCP, MG, GOL |
 |
Crystal Structure Of Acbi4-Mediated Ternary Complex Of Kras G12R Gcp With Pvhl:Elonginc:Elonginb
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2025-11-12 Classification: ONCOPROTEIN Ligands: SO4, A1JHI, GCP, MG |
 |
Organism: Vibrio cholerae
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GCP, GOL, MG |
 |
E.Coli Transcription Translation Coupling Complex In Ttc-B State 4 (Subclass 2) Containing Mrna With 27-Mer Spacer, Nusg, Nusa, Fmet-Trna(Imet), Phe-Trna(Phe), And Gdpcp
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: RIBOSOME Ligands: MG, GCP |
 |
E.Coli Transcription Translation Coupling Complex In Ttc-B State 5 (Subclass 2) Containing Mrna With 27-Mer Spacer, Nusg, Nusa, Fmet-Trna(Imet), Phe-Trna(Phe), And Gdpcp
Organism: Escherichia coli k-12, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-08-13 Classification: RIBOSOME Ligands: MG, GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-B-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-04-30 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-A
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-B
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx:50S-Hflx-C
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Erythromycin:50S-Hflx-A-Ery
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, ERY |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Chloramphenicol:50S-Hflx-B-Clm
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, CLM |
 |
Structure Of Mycobacterium Smegmatis 50S Ribosomal Subunit Bound To Hflx And Chloramphenicol:50S-Hflx-A-Clm
Organism: Mycolicibacterium smegmatis mc2 155
Method: ELECTRON MICROSCOPY Release Date: 2025-02-19 Classification: RIBOSOME Ligands: GCP, CLM |
 |
Structure Of A Yeast 48S-Auc Preinitiation Complex In Swivelled Conformation (Model Py48S-Auc-Swiv-Eif1)
Organism: Saccharomyces cerevisiae s288c, Kluyveromyces lactis, Kluyveromyces lactis nrrl y-1140
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: RIBOSOME Ligands: MG, ZN, GCP, MET |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2024-10-02 Classification: ONCOPROTEIN Ligands: GCP, YLE, MG |

