Search Count: 233
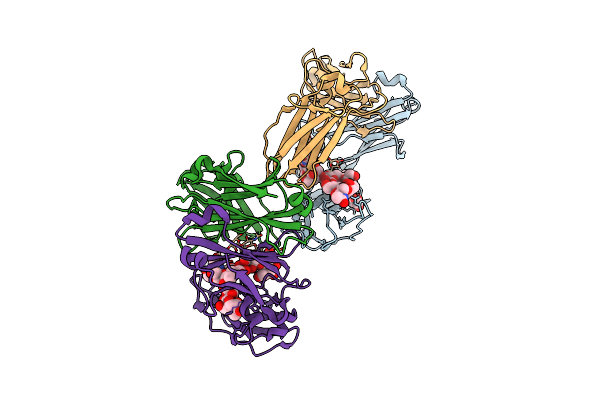 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-10-22 Classification: IMMUNE SYSTEM Ligands: GAL |
 |
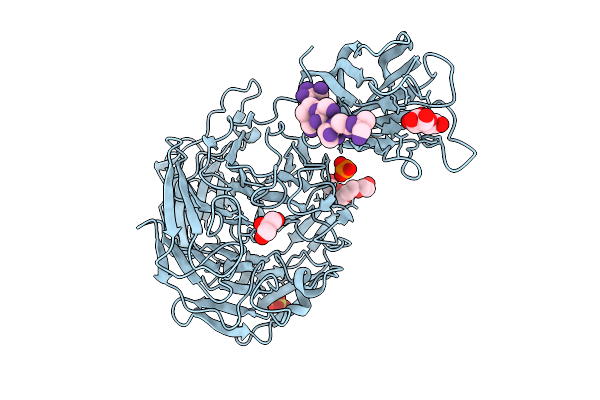
Structure Of S1_8A, A Lambda-Carrageenan Specific Sulfatase, In Complex With Monosaccharide
Organism: Pseudoalteromonas distincta
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA, EDO |
 |
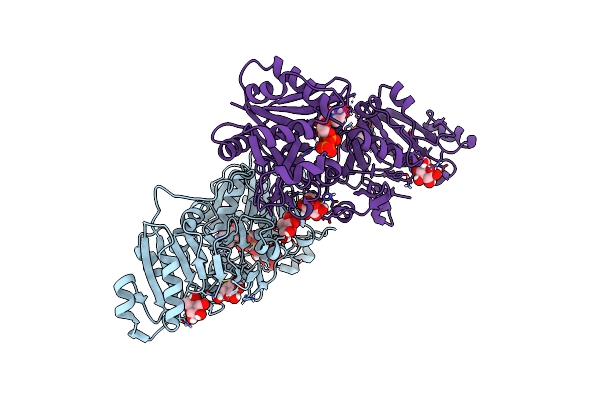
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Release Date: 2025-08-06 Classification: OXIDOREDUCTASE Ligands: GAL, PG4, GOL, PO4, CA, SO4 |
 |
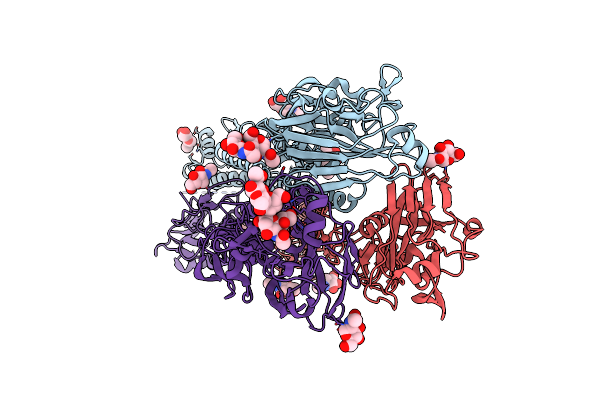
Organism: Acanthamoeba polyphaga mimivirus
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: VIRAL PROTEIN Ligands: UDP, MN, BGC, GAL |
 |
Crystal Structure Of H5 Hemagglutinin From Swan-Infecting H5N8 Influenza Virus In Complex With Lsta
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2025-05-14 Classification: VIRAL PROTEIN/IMMUNE SYSTEM Ligands: NAG, GAL, SIA |
 |
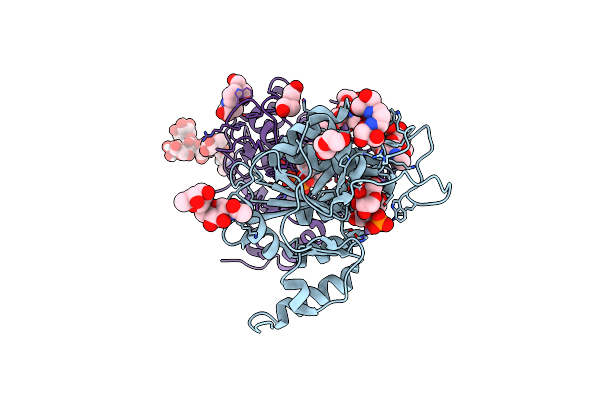
Structure-Based Mechanism And Specificity Of Human Galactosyltransferase B3Galt5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: MN, UDP, B3P, PEG, GAL |
 |
Structure-Based Mechanism And Specificity Of Human Galactosyltransferase B3Galt5
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-04-09 Classification: TRANSFERASE Ligands: MN, UDP, GAL, B3P, PEG |
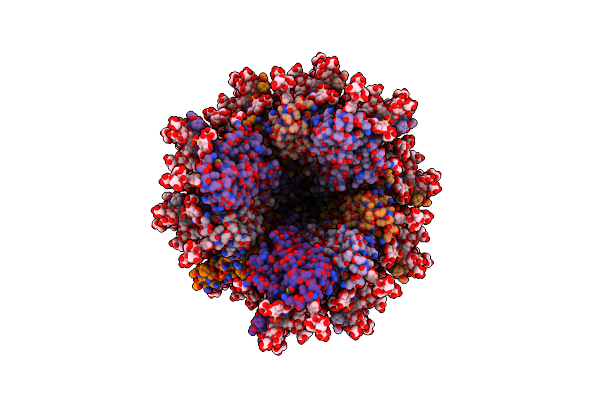 |
Organism: Oxyplasma meridianum
Method: ELECTRON MICROSCOPY Release Date: 2025-01-01 Classification: PROTEIN FIBRIL Ligands: GAL |
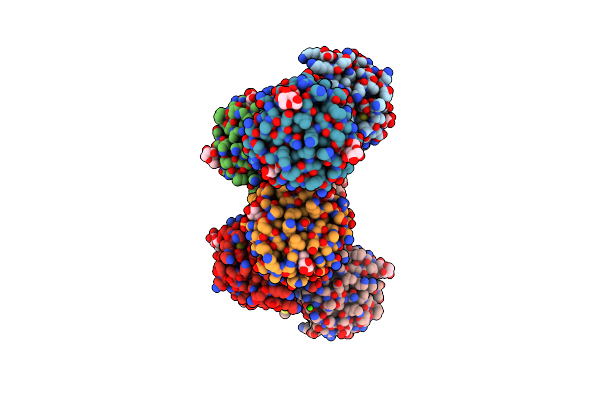 |
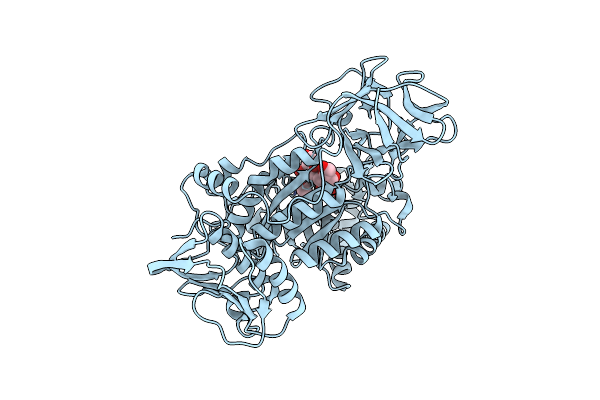
Organism: Mytilus californianus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2024-12-18 Classification: SUGAR BINDING PROTEIN Ligands: GAL, CA |
 |
Crystal Structure Of Sar11_0769 From 'Candidatus Pelagibacter Ubique' Htcc1062 Bound To A Co-Purified Ligand, Beta-Galactopyranose
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: GAL |
 |
Crystal Structure Of Lacto-N-Biosidase Strlnbase From Streptomyces Sp. Strain 142, Lacto-N-Biose Complex
Organism: Streptomyces sp.
Method: X-RAY DIFFRACTION Release Date: 2023-12-27 Classification: HYDROLASE Ligands: GAL, NDG |
 |
Structure Of The Human Cytoplasmic Ribosome With Human Trna Tyr(Galq34) And Mrna(Uau) (Non-Rotated State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: RIBOSOME Ligands: MG, GAL, ZN |
 |
Structure Of The Human Cytoplasmic Ribosome With Human Trna Tyr(Galq34) And Mrna(Uau) (Rotated State)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-12-06 Classification: RIBOSOME Ligands: GAL, MG, ZN |
 |
Structure Of The Bacterial Ribosome With Human Trna Tyr(Galq34) And Mrna(Uau)
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: RIBOSOME Ligands: MG, GAL |
 |
Structure Of The Bacterial Ribosome With Human Trna Tyr(Galq34) And Mrna(Uac)
Organism: Escherichia coli, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-25 Classification: RIBOSOME Ligands: MG, GAL |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 3'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, GAL |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 6'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, EDO, GAL |
 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: MEMBRANE PROTEIN Ligands: GAL |
 |
Crystal Structure Of Flavihumibacter Petaseus Gh31 Alpha-Galactosidase In Complex With Galactose
Organism: Flavihumibacter petaseus nbrc 106054
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: GLA, GAL, EDO, CL |
 |
Crystal Structure Of Flavihumibacter Petaseus Gh31 Alpha-Galactosidase Mutant D304A In Complex With Alpha-1,4-Galactobiose
Organism: Flavihumibacter petaseus nbrc 106054
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-07-26 Classification: HYDROLASE Ligands: GLA, GAL, EDO, CL |

