Search Count: 98
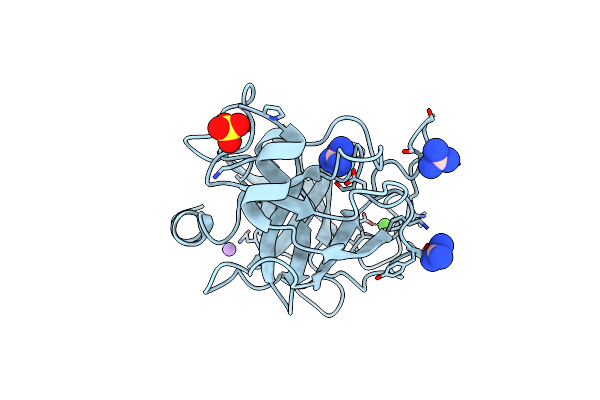 |
Crystal Structure Of Trypsin-Guanidine Complex At 2.05 Angstroms Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-18 Classification: HYDROLASE Ligands: CA, NA, SO4, CL, GAI |
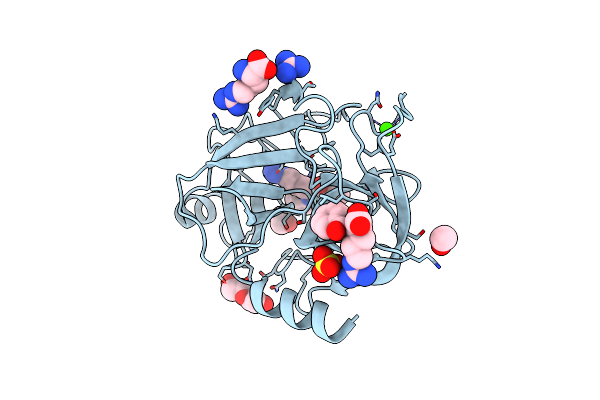 |
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-08-14 Classification: HYDROLASE Ligands: I4Q, JNR, ARG, GAI, SO4, CA, CL, PEG, EOH |
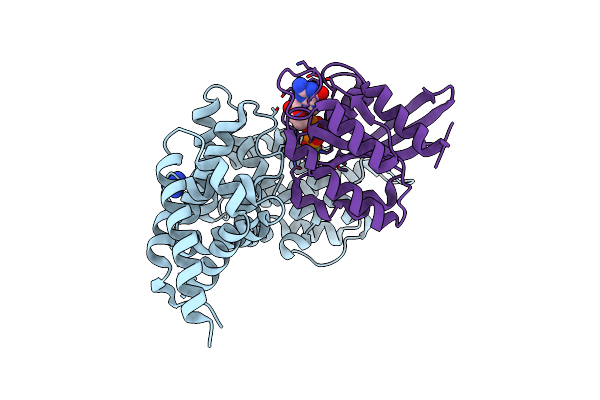 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-11-29 Classification: HYDROLASE Ligands: GDP, MGF, MG, GAI |
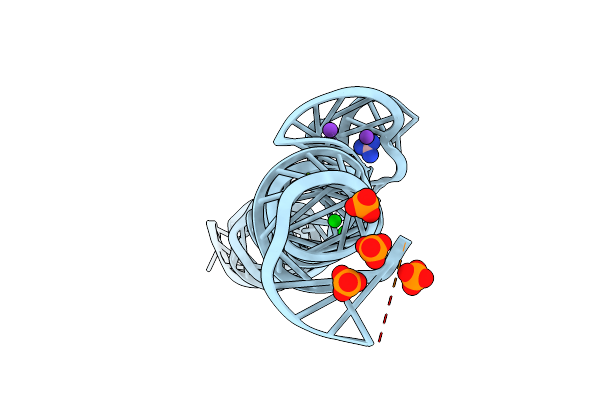 |
Organism: Burkholderia sp. tji49
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-01-12 Classification: RNA Ligands: GAI, PO4, SR, MG, K |
 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-617
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2021-08-25 Classification: HYDROLASE Ligands: QWT, ZN, MG, PEG, GOL, GAI |
 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-656
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-08-25 Classification: HYDROLASE Ligands: ZN, MG, FMT, GAI, GOL, QWZ |
 |
Structure Of Recombinant Human Beta-Glucocerebrosidase In Complex With Bodipy Functionalised Epoxide Activity Based Probe
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-06-30 Classification: HYDROLASE Ligands: NAG, Q65, K35, SO4, EDO, GOL, GAI, ACT |
 |
The Structure Of Agmatinase From E. Coli At 3.2 A Displaying Guanidine In The Active Site
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2021-05-12 Classification: HYDROLASE Ligands: MN, GAI |
 |
Organism: Bacteroides fluxus yit 12057
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2021-02-17 Classification: SUGAR BINDING PROTEIN Ligands: GAI, PEG |
 |
Surface Glycan-Binding Protein B From Bacteroides Fluxus In Complex With Mixed-Linkage Glucotriose
Organism: Bacteroides fluxus yit 12057
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-02-17 Classification: SUGAR BINDING PROTEIN Ligands: GAI |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2020-12-30 Classification: RNA Ligands: GAI, CA, GOL |
 |
Organism: Acanthamoeba castellanii str. neff
Method: X-RAY DIFFRACTION Resolution:2.92 Å Release Date: 2020-10-21 Classification: OXIDOREDUCTASE Ligands: HEM, CL6, GAI |
 |
Organism: Thermotoga maritima msb8
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-07-15 Classification: TRANSPORT PROTEIN Ligands: GAI, EDO |
 |
Crystal Structure Of Ketosteroid Isomerase From Mycobacterium Hassiacum (Mhksi)
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: SO4, GAI |
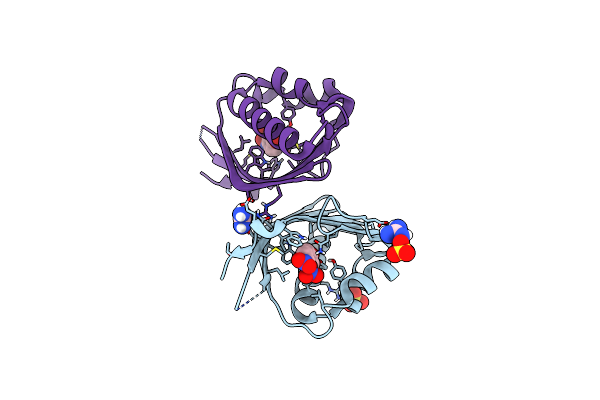 |
Crystal Structure Of Ketosteroid Isomerase D38N Mutant From Mycobacterium Hassiacum (Mhksi) Bound To 3,4-Dinitrophenol
Organism: Mycobacterium hassiacum (strain dsm 44199 / cip 105218 / jcm 12690 / 3849)
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2020-05-27 Classification: ISOMERASE Ligands: DNX, SO4, GAI |
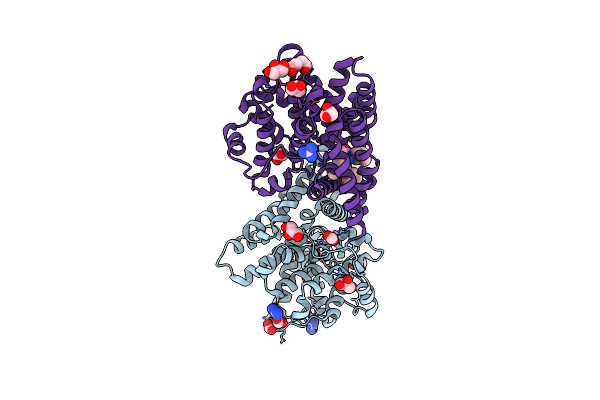 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-0769
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2020-02-05 Classification: HYDROLASE Ligands: ZN, MG, FMT, GAI, GOL, PEG, J2E |
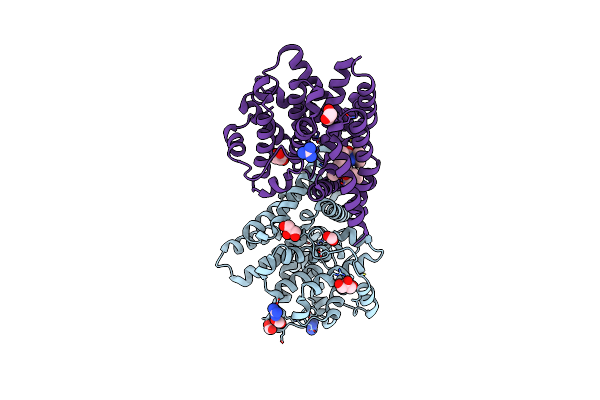 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-1361
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2020-02-05 Classification: HYDROLASE Ligands: ZN, MG, FMT, GAI, GOL, 863 |
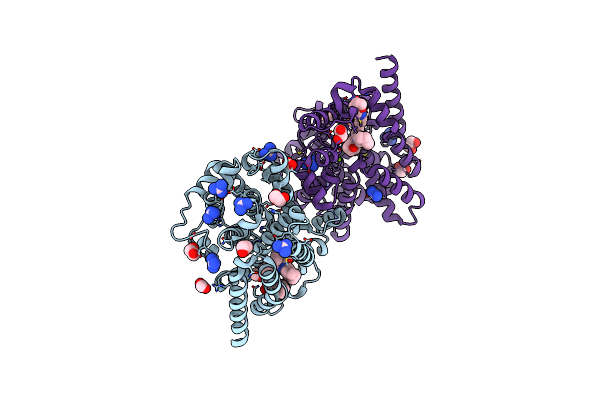 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-053
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: MG, ZN, GAI, EDO, FMT, JX2 |
 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-1018
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: ZN, MG, FMT, GAI, GOL, K3W |
 |
Crystal Structure Of T. Brucei Pde-B1 Catalytic Domain With Inhibitor Npd-1039
Organism: Trypanosoma brucei
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2019-07-24 Classification: HYDROLASE Ligands: ZN, MG, FMT, GAI, GOL, K1Q |

