Search Count: 25
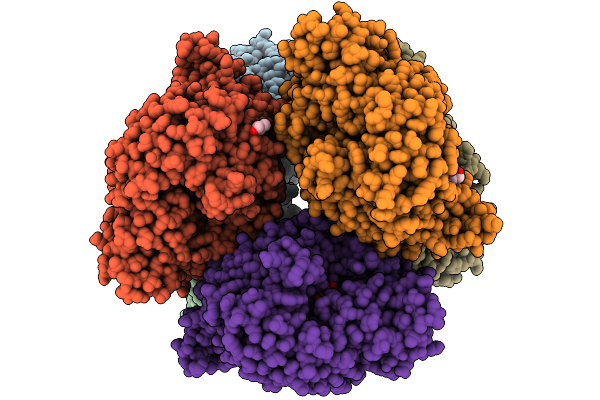 |
Structural Basis Of The Bifunctionality Of M. Salinexigens Zyf650T Glucosylglycerol Phosphorylase In Glucosylglycerol Catabolism
Organism: Marinobacter salinexigens
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2025-09-10 Classification: STRUCTURAL PROTEIN Ligands: GLC, GOL, NA, G1P |
 |
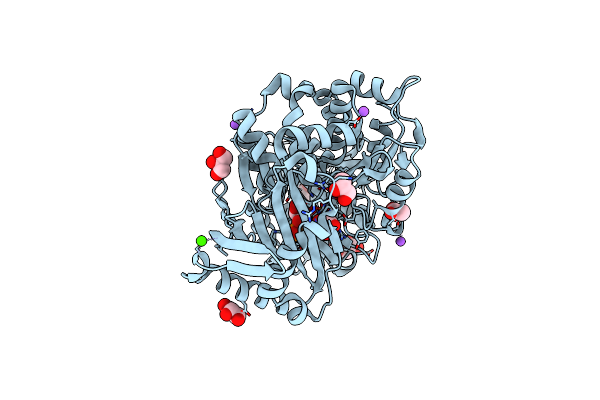
Crystal Structure Of Bifunctional Glmu From Staphylococcus Aureus Nctc 8325 Complexed With Utp, Coa And Glc 1-P
Organism: Staphylococcus aureus subsp. aureus nctc 8325
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-05-14 Classification: TRANSFERASE Ligands: COA, MG, G1P, UTP |
 |
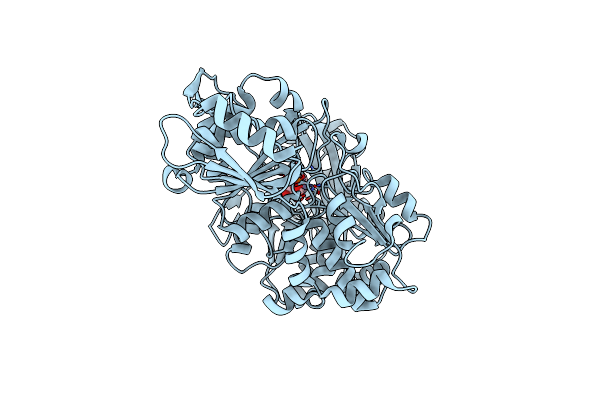
Structure Of Baltic Herring (Clupea Harengus) Phosphoglucomutase 5 (Pgm5) With Bound Glucose-1-Phosphate
Organism: Clupea harengus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-12-16 Classification: ISOMERASE Ligands: NI, CA, G1P, GOL, ACT, NA |
 |
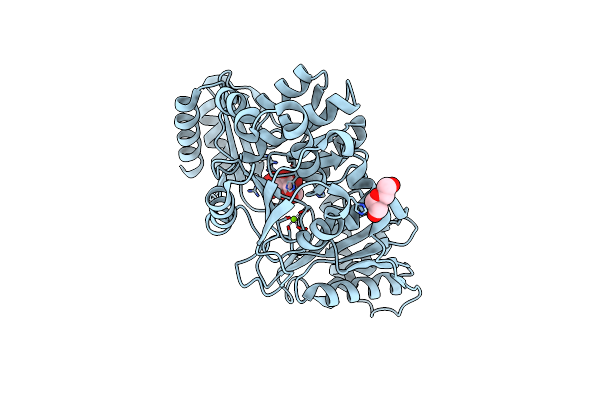
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2020-04-08 Classification: ISOMERASE Ligands: ZN, G1P |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: MG, PEG, G1P |
 |
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: CA, G1P, PEG |
 |
Paenibacillus Sp. Ym1 Laminaribiose Phosphorylase With Alpha-Glc-1-Phosphate Bound
Organism: Paenibacillus sp. ym1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-13 Classification: HYDROLASE Ligands: SO4, G1P, EDO, CL |
 |
Organism: Mycobacterium tuberculosis (strain atcc 25177 / h37ra)
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-05-02 Classification: TRANSFERASE Ligands: MG, G1P, ACO, PEG, EDO |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Alpha-D-Glucose-1-Phosphate
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: G1P, GLC |
 |
Crystal Structure Of Phosphoglucomutase From Xanthomonas Citri Complexed With Glucose-1-Phosphate
Organism: Xanthomonas axonopodis pv. citri (strain 306)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-06-01 Classification: ISOMERASE Ligands: G1P, MG |
 |
Crystal Structure Of Glucose-1-Phosphate Bound Nucleotidylated Human Galactose-1-Phosphate Uridylyltransferase
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2016-03-30 Classification: TRANSFERASE Ligands: ZN, H2U, EDO, G1P |
 |
Structure Of The Rbp Of Lactococcal Phage 1358 In Complex With Glucose-1-Phosphate
Organism: Lactococcus phage 1358
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2014-04-30 Classification: VIRAL PROTEIN Ligands: G1P |
 |
Crystal Structure Of Glucose 1-Phosphate Thymidylyltransferase From Aneurinibacillus Thermoaerophilus Complexed With Thymidine And Glucose-1-Phosphate
Organism: Aneurinibacillus thermoaerophilus
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-10-23 Classification: TRANSFERASE Ligands: G1P, THM, SO4 |
 |
Structure Of Beta-Phosphoglucomutase Inhibited With Glucose-6- Phosphate, Glucose-1-Phosphate And Beryllium Trifluoride
Organism: Lactococcus lactis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2010-05-26 Classification: ISOMERASE Ligands: BEF, MG, G1P, BG6, NA, PEG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2008-09-09 Classification: ISOMERASE Ligands: G1P, ZN |
 |
Crystal Structure Of Sphingomonas Elodea Atcc 31461 Glucose-1- Phosphate Uridylyltransferase In Complex With Glucose-1-Phosphate.
Organism: Sphingomonas elodea
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2007-05-22 Classification: TRANSFERASE Ligands: G1P |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2005-11-22 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: G1P, MG |
 |
Structural Basis For Allosteric Regulation And Substrate Specificity Of The Non-Phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase (Gapn) From Thermoproteus Tenax
Organism: Thermoproteus tenax
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2004-08-05 Classification: OXIDOREDUCTASE Ligands: NAP, G1P, NA |
 |
Structural Basis For Allosteric Regulation And Substrate Specificity Of The Non-Phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase (Gapn) From Thermoproteus Tenax
Organism: Thermoproteus tenax
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-08-05 Classification: OXIDOREDUCTASE Ligands: G1P, NAD, NA |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-01-20 Classification: ISOMERASE Ligands: G1P, ZN |

