Search Count: 18
 |
Crystal Structure Of Mox-1 Complexed With A Boronic Acid Transition State Inhibitor S02030
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-06-26 Classification: HYDROLASE Ligands: ZXM, ACT, ZN |
 |
Organism: Vitis vinifera
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2018-10-03 Classification: HYDROLASE INHIBITOR Ligands: EDO |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: MG, PEG |
 |
The Ternary Structure Of D-Lactate Dehydrogenase From Pseudomonas Aeruginosa With Nadh And Oxamate
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: FMT, NAI, OXM, PGE, PEG, PG4 |
 |
The Ternary Structure Of D-Lactate Dehydrogenase From Fusobacterium Nucleatum With Nadh And Oxamate
Organism: Fusobacterium nucleatum subsp. nucleatum (strain atcc 25586 / cip 101130 / jcm 8532 / lmg 13131)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: NAI, OXM |
 |
Organism: Fusobacterium nucleatum subsp. nucleatum (strain atcc 25586 / cip 101130 / jcm 8532 / lmg 13131)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: SO4, GOL |
 |
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2018-09-19 Classification: OXIDOREDUCTASE Ligands: ACT |
 |
The Ternary Structure Of D-Mandelate Dehydrogenase With Nadh And Anilino(Oxo)Acetate
Organism: Enterococcus faecium do
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2017-04-05 Classification: OXIDOREDUCTASE Ligands: NAD, AOT, GOL, SO4 |
 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: GOL, CA, PEG |
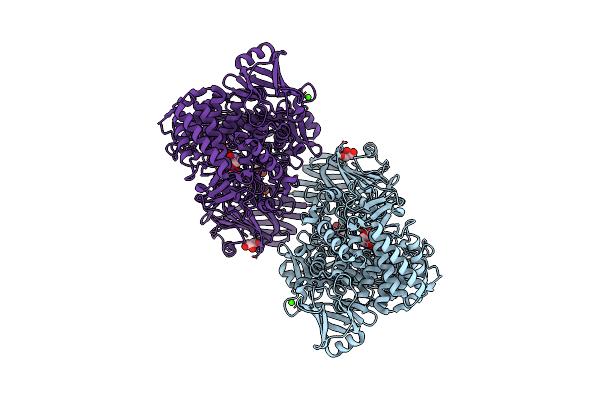 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Sophorose
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: GOL, CA |
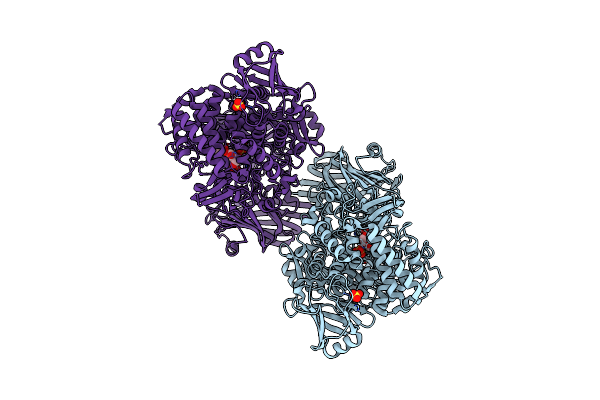 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Sophorose, Isofagomine, Sulfate Ion
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: IFM, SO4 |
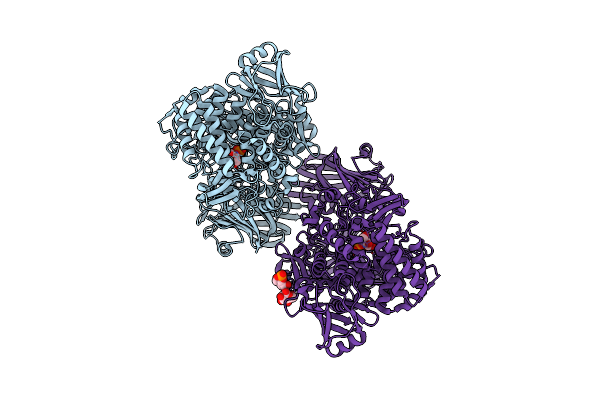 |
Crystal Structure Of 1,2-Beta-Oligoglucan Phosphorylase From Lachnoclostridium Phytofermentans In Complex With Alpha-D-Glucose-1-Phosphate
Organism: Clostridium phytofermentans isdg
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2017-03-01 Classification: TRANSFERASE Ligands: G1P, GLC |
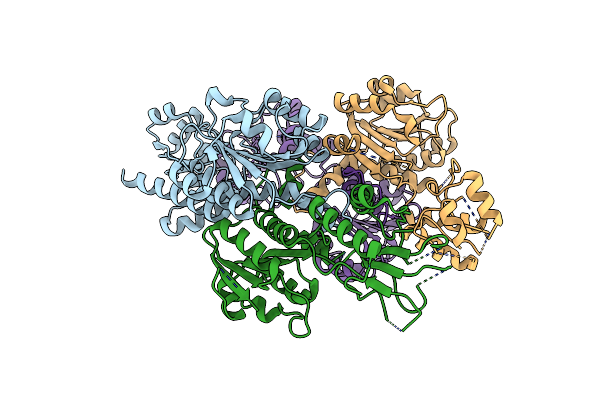 |
Organism: Escherichia coli bl21(de3)
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2015-07-08 Classification: OXIDOREDUCTASE |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE |
 |
Organism: Enterococcus faecium
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2014-07-23 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: GOL |
 |
L-Lactate Dehydrogenase From Thermus Caldophilus Gk24 Complexed With Oxamate, Nadh And Fbp
Organism: Thermus caldophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-03-06 Classification: OXIDOREDUCTASE Ligands: OXM, FBP, NAD, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2009-11-03 Classification: TRANSCRIPTION Ligands: Z27 |

