Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
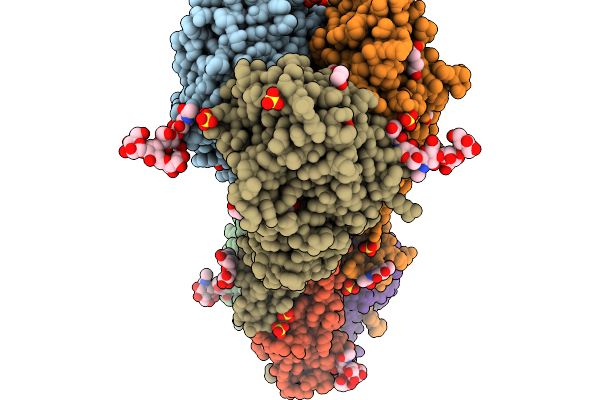 |
Crystal Structure Of The H5 Influenza Virus Hemagglutinin From A/Duck/France/161108H/2016 (H5N8) Clade 2.3.4.4B
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: VIRAL PROTEIN Ligands: EDO, SO4, NAG, PEG |
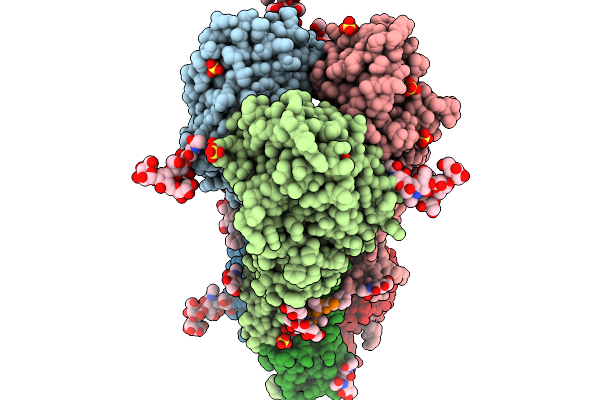 |
Crystal Structure Of The H5 Influenza Virus Hemagglutinin From A/Duck/France/161108H/2016 (H5N8) Clade 2.3.4.4B In Complex With O-Linked Glycan 26
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: VIRAL PROTEIN Ligands: NAG, GOL, SO4 |
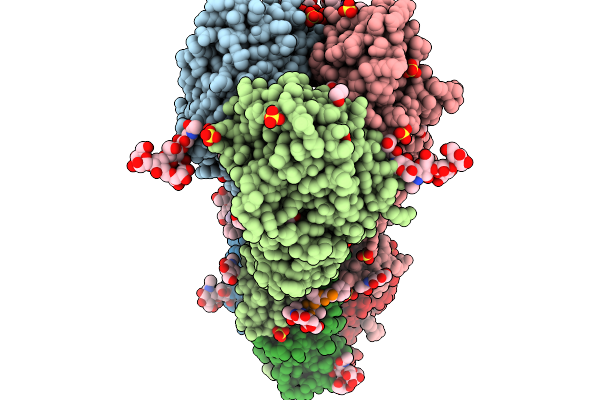 |
Crystal Structure Of The H5 Influenza Virus Hemagglutinin From A/Duck/France/161108H/2016 (H5N8) Clade 2.3.4.4B In Complex With O-Linked Glycan 25
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: VIRAL PROTEIN Ligands: EDO, SO4, NAG |
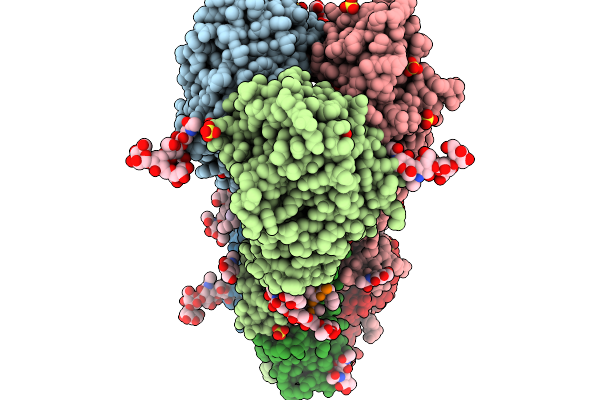 |
Crystal Structure Of The H5 Influenza Virus Hemagglutinin From A/Duck/France/161108H/2016 (H5N8) Clade 2.3.4.4B In Complex With O-Linked Glycan 7
Organism: Influenza a virus, Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: VIRAL PROTEIN Ligands: NAG, GOL, SO4 |
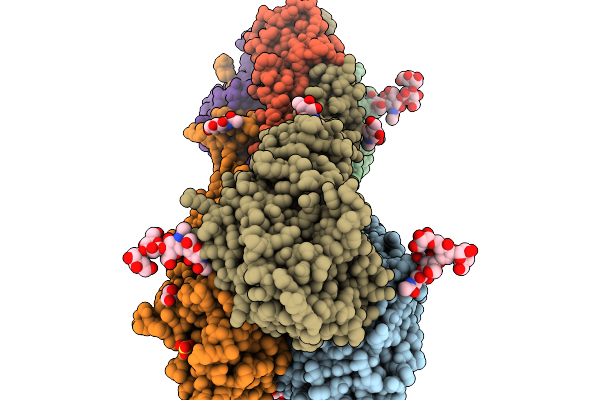 |
Crystal Structure Of The H5 Influenza Virus Hemagglutinin From A/Duck/France/161108H/2016 (H5N8) Clade 2.3.4.4B In Complex With Avian Receptor Analog Lsta
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Release Date: 2026-01-14 Classification: VIRAL PROTEIN Ligands: NAG, GOL, SO4, CIT |
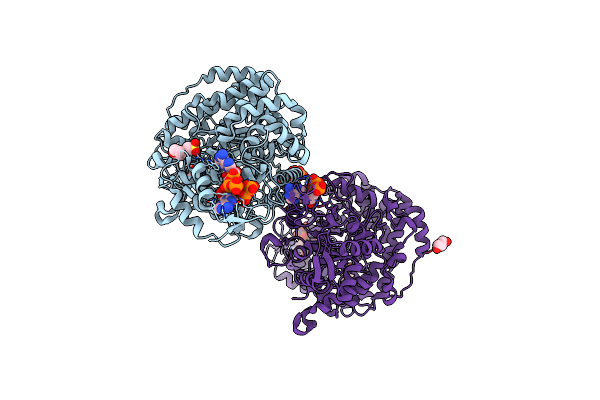 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit Bound To Two Atp Molecules
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.62 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: ATP, MG, GOL, MES |
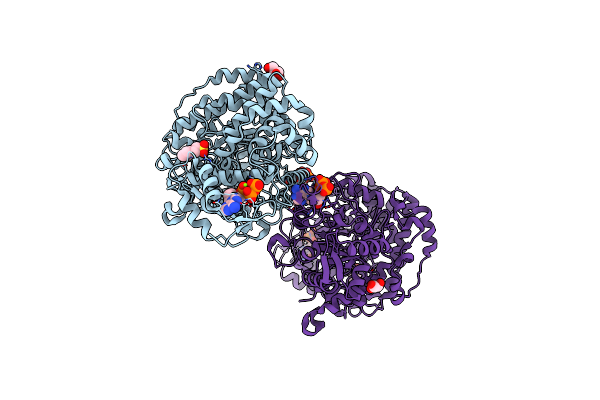 |
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit Bound To Datp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, MES, GOL |
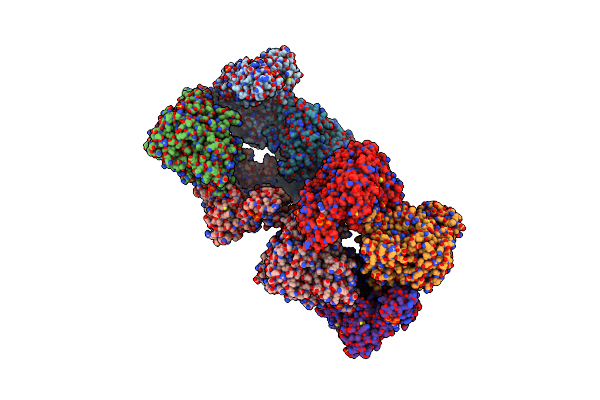 |
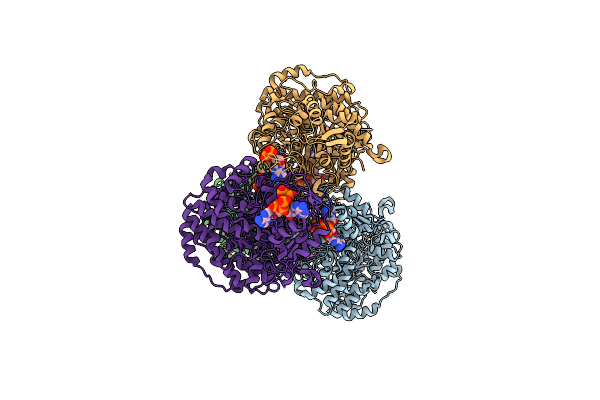
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Cdp And Two Molecules Of Atp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: SO4, ATP, MG, CDP |
 |
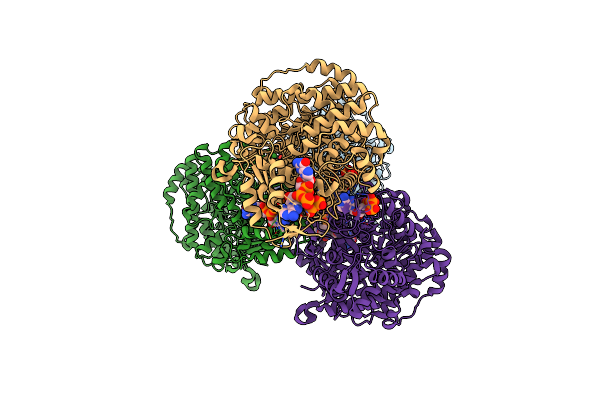
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Datp And Atp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, ATP |
 |
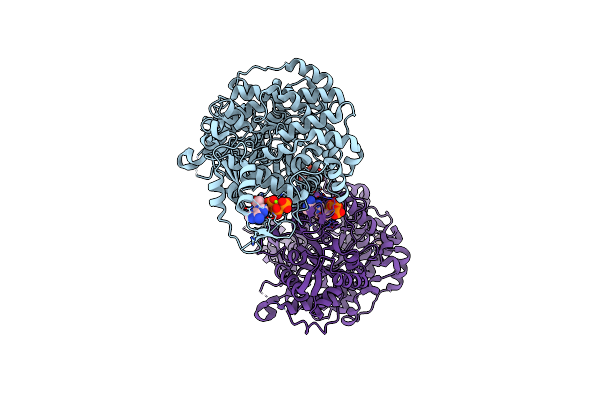
Crystal Structure Of E. Coli Class Ia Ribonucleotide Reductase Alpha Subunit W28A Variant Bound To Datp And Gtp
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, GTP |
 |
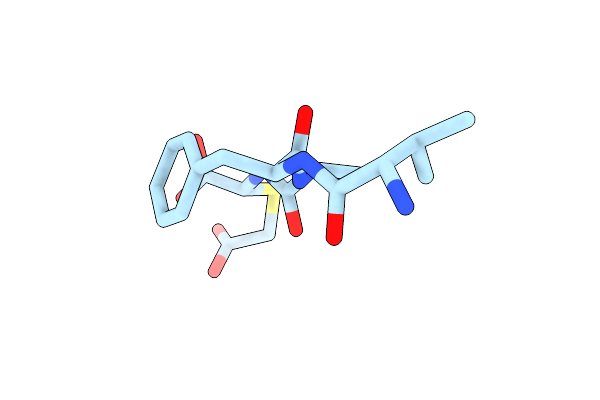
Crystal Structure Of Datp Bound E. Coli Class Ia Ribonucleotide Reductase Alpha Construct Fused With The C-Terminal Tail Of E. Coli Class Ia Beta Subunit
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-09-04 Classification: OXIDOREDUCTASE Ligands: DTP, MG, NA, CL, GOL |
 |
Organism: Pseudomonas syringae
Method: ELECTRON CRYSTALLOGRAPHY Resolution:1.00 Å Release Date: 2019-08-07 Classification: UNKNOWN FUNCTION Ligands: HOH |
 |
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:2.36 Å Release Date: 2019-01-16 Classification: LYASE Ligands: BTL |
 |
Organism: Desulfovibrio alaskensis (strain g20)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE Ligands: CHT, GOL |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-09-28 Classification: LYASE |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2016-09-28 Classification: LYASE |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE |
 |
Organism: Desulfovibrio alaskensis
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2016-09-28 Classification: LYASE |
 |
Organism: Escherichia coli o157:h7
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-07-13 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |
 |
Organism: Escherichia coli 1303
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-06-22 Classification: OXIDOREDUCTASE Ligands: FEO, SO4 |