Search Count: 101
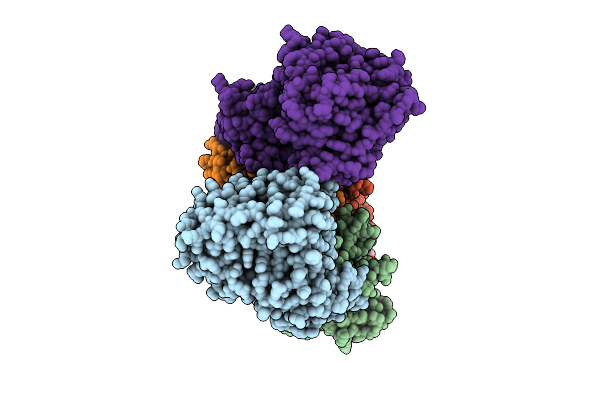 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Middle State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.75 Å Release Date: 2025-08-06 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, LMT, FES, FAD |
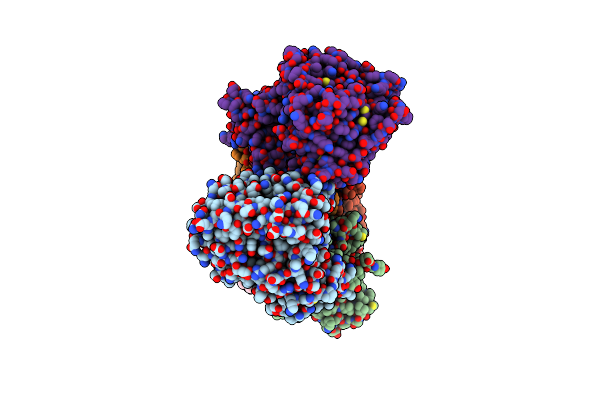 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
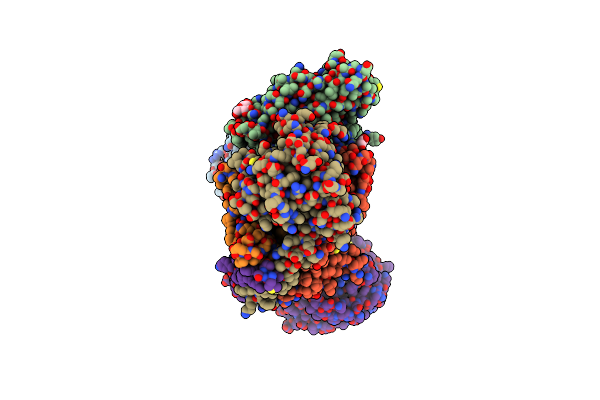 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrc-T225Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.59 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
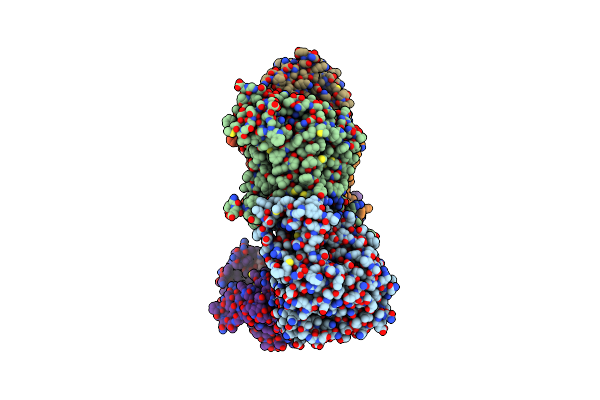 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, FMN, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-T236Y Mutant From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.31 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: RBF, LMT, CA, FMN, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Upper State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.65 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, In The Absence Of Na+, Down State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.11 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Stable State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.61 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae Reduced By Nadh, With Bound Korormicin A, Shifted State
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.93 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, PEE, LMT, IQT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh, With Bound Aurachin D-42
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:3.18 Å Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, 0NI, PEE, LMT, CA, FES, FAD |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase From Vibrio Cholerae Reduced By Nadh
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, CA, FES, FAD, NAI |
 |
Cryo-Em Structure Of Na+-Translocating Nadh-Ubiquinone Oxidoreductase Nqrb-G141A Mutant From Vibrio Cholerae With Bound Korormicin A
Organism: Vibrio cholerae o395
Method: ELECTRON MICROSCOPY Resolution:2.68 Å Release Date: 2025-04-23 Classification: MEMBRANE PROTEIN Ligands: FMN, RBF, LMT, PEE, IQT, CA, FES, FAD |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Resolution:3.19 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: 9UJ, A1LYC |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.36 Å Release Date: 2025-04-02 Classification: MEMBRANE PROTEIN Ligands: A1LYD |
 |
Serial Femtosecond Crystallography Structure Of Co Bound Ba3- Type Cytochrome C Oxidase Without Pump Laser Irradiation
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
 |
Serial Femtosecond Crystallography Structure Of Photo Dissociated Co From Ba3- Type Cytochrome C Oxidase Determined By Extrapolation Method
Organism: Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-15 Classification: ELECTRON TRANSPORT Ligands: CU, HEM, HAS, OLC, CMO, CUA |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.56 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr2 In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.53 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |
 |
Cryo-Em Structure Of The Potassium-Selective Channelrhodopsin Hckcr1 H225F Mutant In Lipid Nanodisc
Organism: Hyphochytrium catenoides
Method: ELECTRON MICROSCOPY Resolution:2.66 Å Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: RET, PSC, PLM |

