Search Count: 16
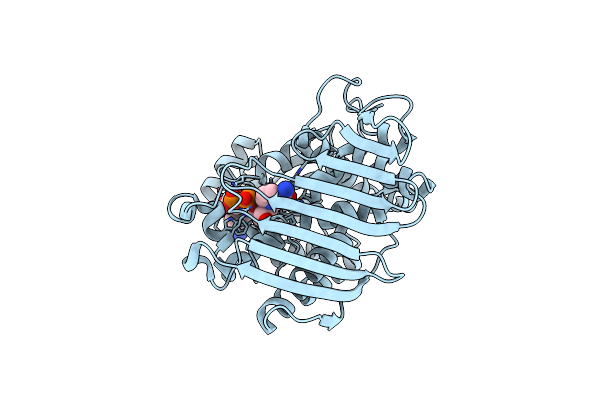 |
Crystal Structure Of Inositol Dehydrogenase Homolog Complexed With Nad+ From Azotobacter Vinelandii
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NAD |
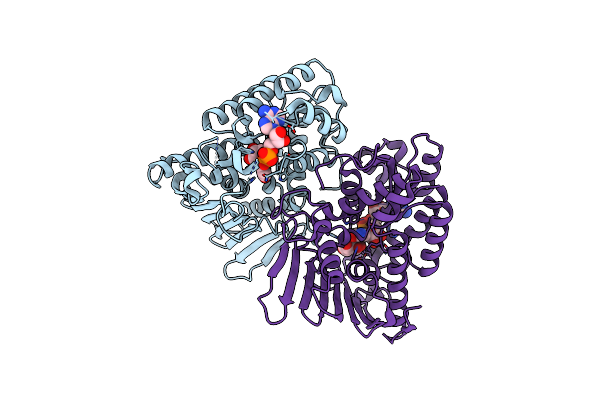 |
Crystal Structure Of Inositol Dehydrogenase Homolog Complexed With Nadh And Myo-Inositol From Azotobacter Vinelandii
Organism: Azotobacter vinelandii (strain dj / atcc baa-1303)
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: NAI, INS |
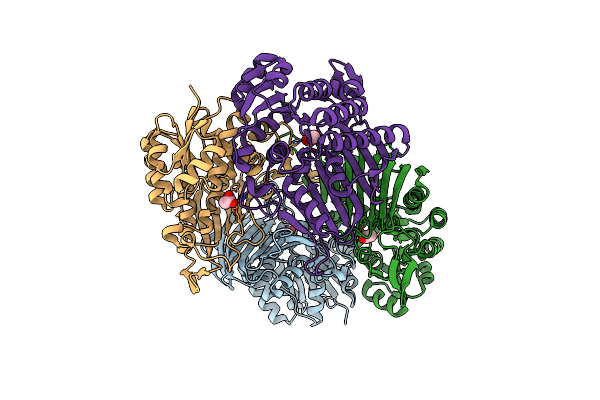 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Apo-Form, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: ACT |
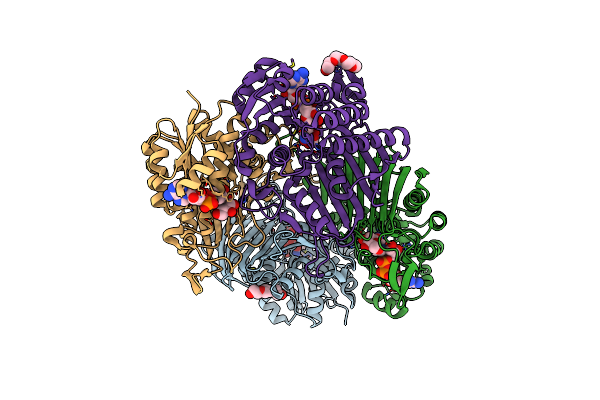 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Complexed With Nadh And L-Glucono-1,5-Lactone, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: NAI, PG4, 8S0 |
 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase R178A Mutant, Complexed With Nad And Myo-Inositol, From Paracoccus Laeviglucosivorans
Organism: Paracoccus laeviglucosivorans
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-12-25 Classification: OXIDOREDUCTASE Ligands: ACT, NAD, INS |
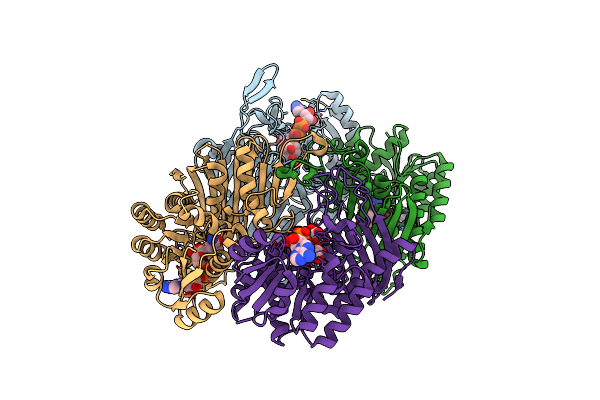 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase With L-Glucose Dehydrogenase Activity Complexed With Myo-Inositol
Organism: Paracoccus laeviglucosivorans nakamura 2015
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: NAD, INS |
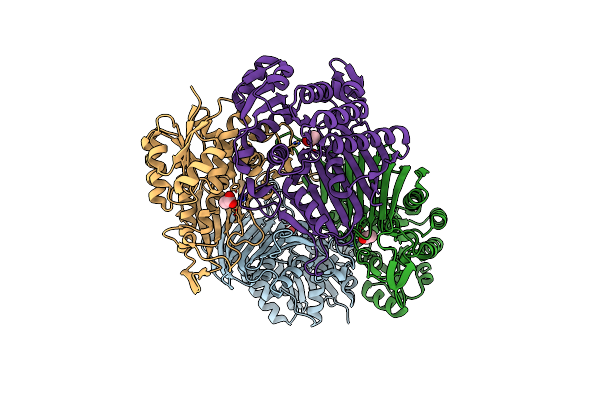 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase With L-Glucose Dehydrogenase Activity
Organism: Paracoccus laeviglucosivorans nakamura 2015
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: ACT |
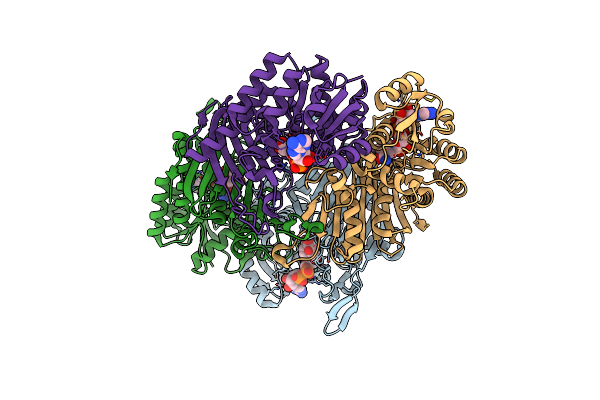 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase With L-Glucose Dehydrogenase Activity Complexed With L-Glucono-1,5-Lactone
Organism: Paracoccus laeviglucosivorans nakamura 2015
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: NAI, 8S0 |
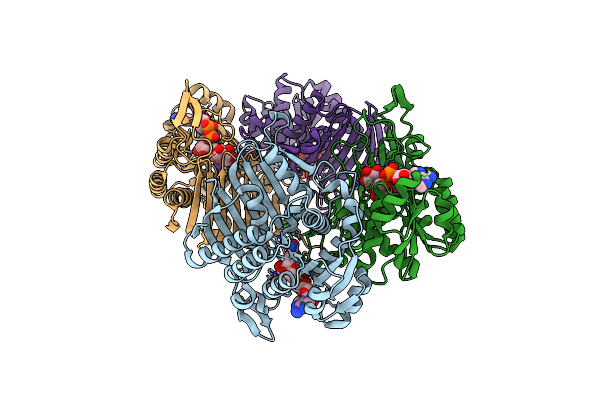 |
Crystal Structure Of Scyllo-Inositol Dehydrogenase With L-Glucose Dehydrogenase Activity Complexed With Scyllo-Inosose
Organism: Paracoccus laeviglucosivorans nakamura 2015
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2018-05-23 Classification: OXIDOREDUCTASE Ligands: NAD, ISE |
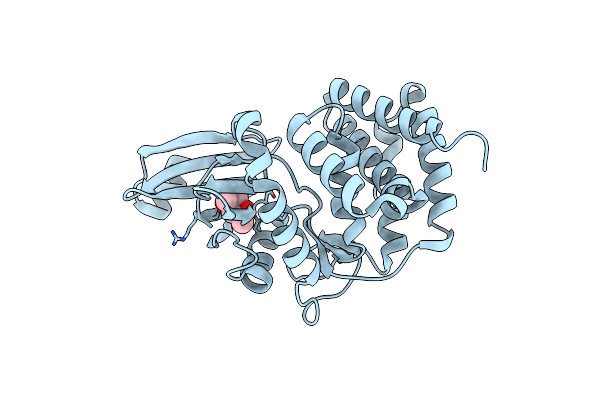 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia, Apo Form
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-07 Classification: TRANSFERASE Ligands: MPD |
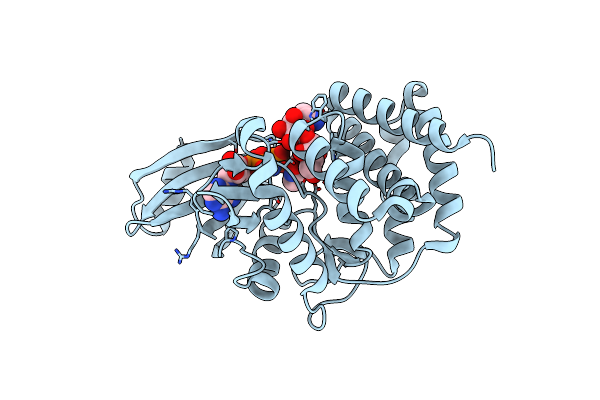 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia, Ternary Complex With Amp-Pnp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ANP, HY0 |
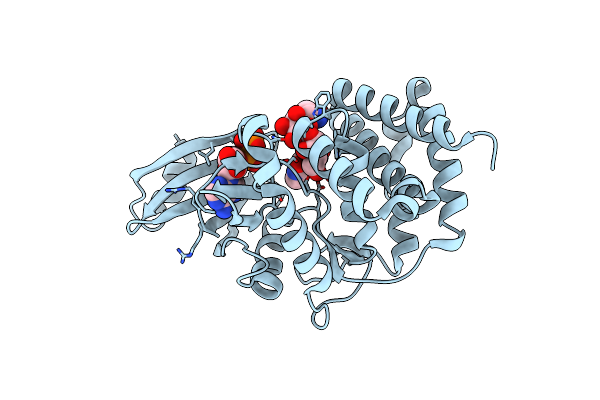 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia, Ternary Complex With Adp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ADP, HY0 |
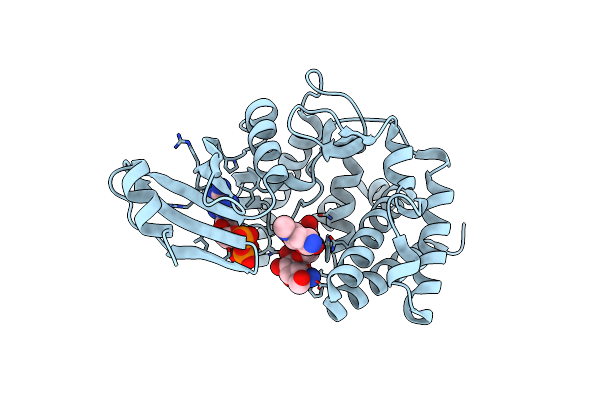 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia (D198A), Ternary Complex With Adp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ADP, HY0 |
 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia (N203A), Ternary Complex With Amp-Pnp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ANP, HY0 |
 |
Crystal Structure Of A Thermostable Mutant Of Aminoglycoside Phosphotransferase Aph(4)-Ia (N202A), Ternary Complex With Amp-Pnp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: ANP, HY0 |
 |
Crystal Structure Of Aminoglycoside Phosphotransferase Aph(4)-Ia, Ternary Complex With Amp-Pnp And Hygromycin B
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2013-08-07 Classification: TRANSFERASE/ANTIBIOTIC Ligands: HY0, ANP, MG |

