Search Count: 15
 |
Organism: Microchaete diplosiphon
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-05-15 Classification: SIGNALING PROTEIN Ligands: CYC |
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2020-11-18 Classification: MEMBRANE PROTEIN Ligands: P2E |
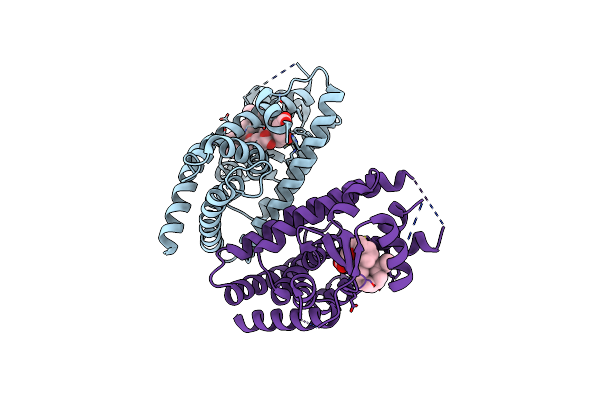 |
Covalent Bond Formation Between Ynone Moiety Of Synthetic Fatty Acid And Hpparg-Lbd
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-02-12 Classification: NUCLEAR PROTEIN Ligands: EJL |
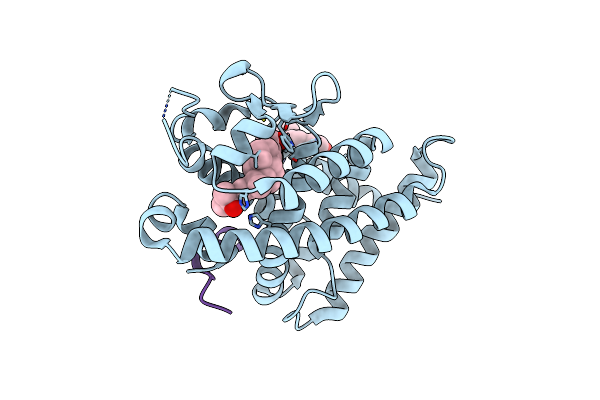 |
Covalent Labeling Of Rvdr-Lbd By Turn-On Fluorescent Probe Mediated By Conjugate Addition And Cyclization
Organism: Rattus norvegicus, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-12 Classification: HORMONE Ligands: YSV, EJO |
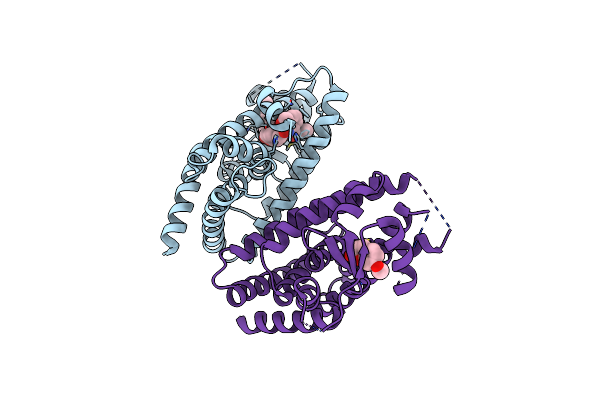 |
Covalent Labeling Of Hpparg-Lbd By Turn-On Fluorescent Probe Mediated By Conjugate Addition And Cyclization
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2020-02-12 Classification: NUCLEAR PROTEIN Ligands: EB6, EBF |
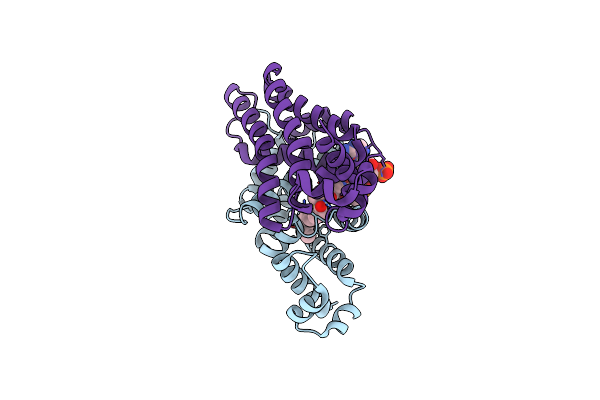 |
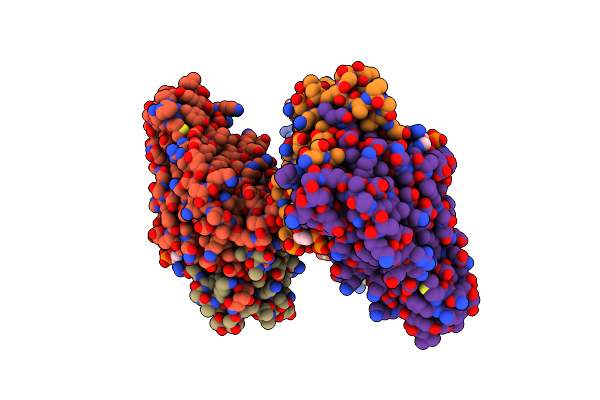
Crystal Structure Of Fadr From Bacillus Subtilis, A Transcriptional Regulator Involved In The Regulation Of Fatty Acid Degradation
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: DCC |
 |
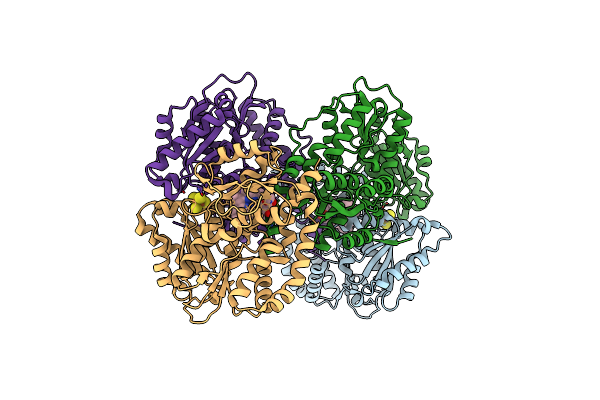
Crystal Structure Of A Transcriptional Regulator Fadr From Bacillus Subtilis In Complex With Stearoyl-Coa
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: ST9 |
 |
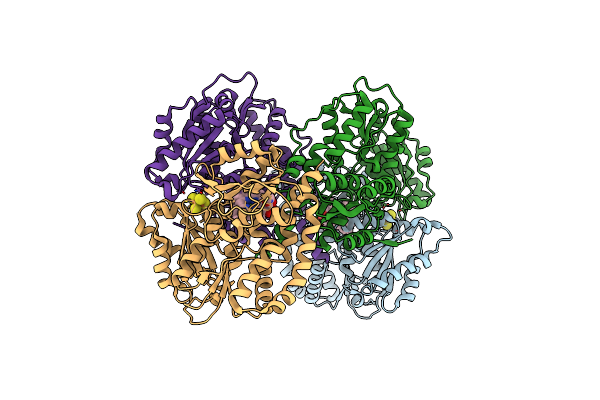
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
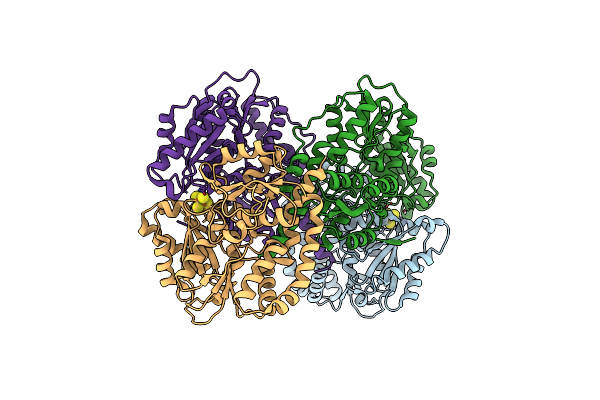
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4, PMR |
 |
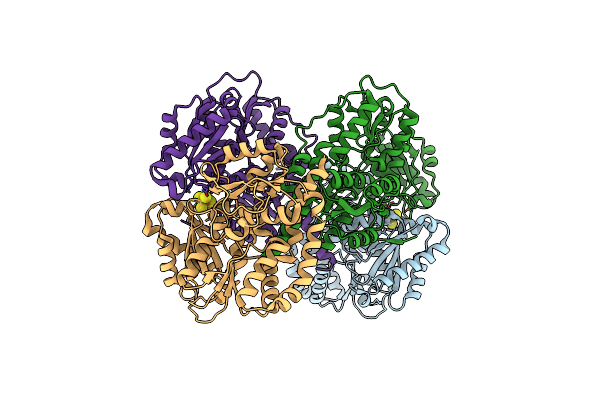
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
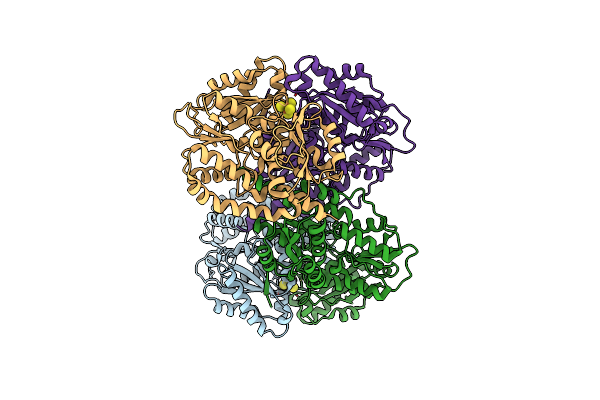
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
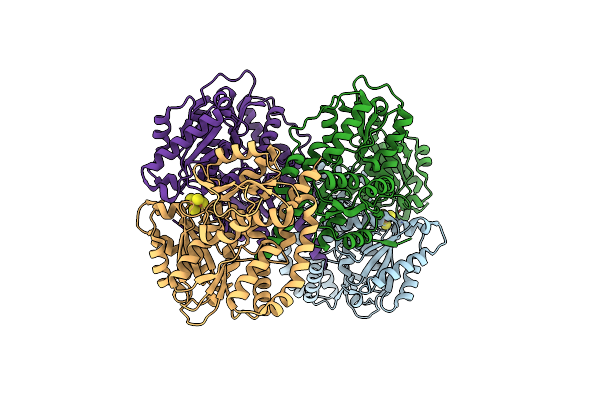
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.91 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
Structure Of The Light-Independent Protochlorophyllide Reductase Catalyzing A Key Reduction For Greening In The Dark
Organism: Rhodobacter capsulatus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: SF4 |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-11-03 Classification: HYDROLASE/Hormone Receptor Ligands: A8S |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2009-11-03 Classification: Hormone Receptor Ligands: A8S |

