Search Count: 65
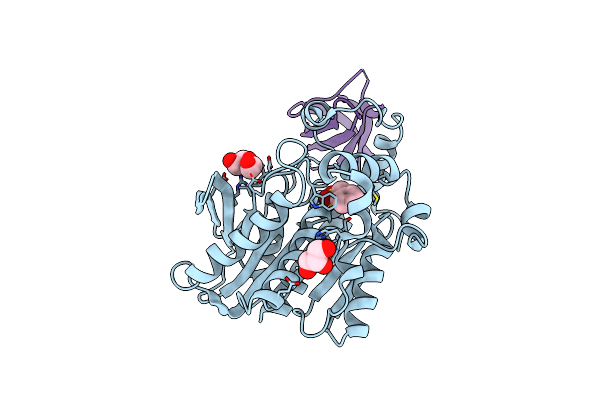 |
Crystal Structure Of The Thermotoga Maritima Cold Shock Protein Mutant Est#13 In Complex With Aacest
Organism: Alicyclobacillus acidocaldarius, Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-15 Classification: PROTEIN BINDING Ligands: PMS, PEG, GOL |
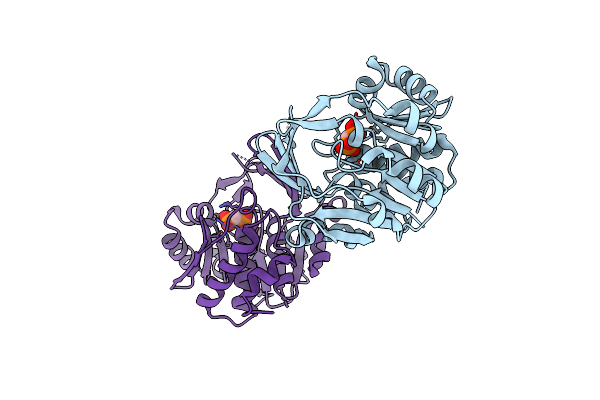 |
Conversion Of Pyrophosphate-Dependent Myo-Inositol-1 Kinase Into Myo-Inositol-3 Kinase By N78L/S89L Mutation
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2021-05-05 Classification: TRANSFERASE Ligands: MDN, MG |
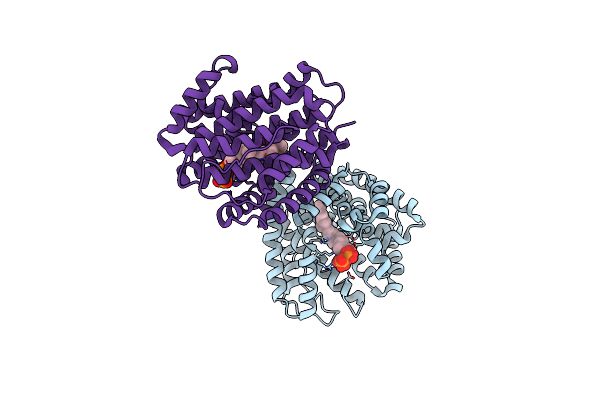 |
Crystal Structure Of Class Ib Terpene Synthase Bound With Geranylcitronellyl Diphosphate
Organism: Bacillus alcalophilus atcc 27647 = cgmcc 1.3604
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-11-18 Classification: LYASE Ligands: ELR, ELU |
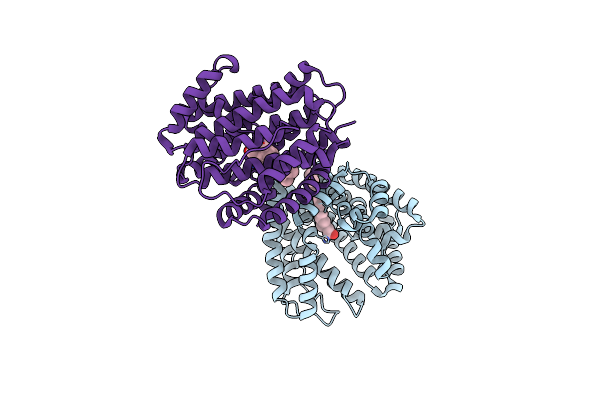 |
Organism: Bacillus alcalophilus atcc 27647 = cgmcc 1.3604
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-11-18 Classification: LYASE Ligands: ELX |
 |
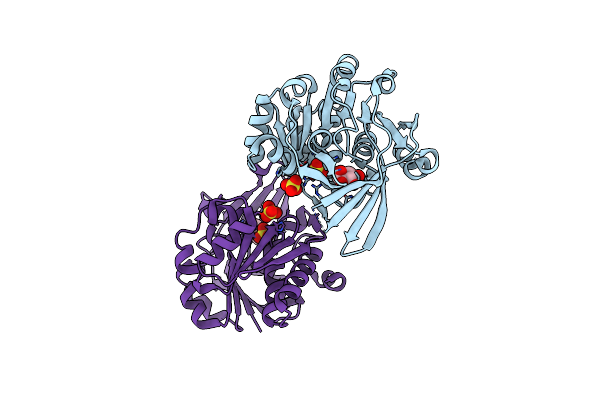
Pyrophosphate-Dependent Kinase In The Ribokinase Family Complexed With A Pyrophosphate Analog And Myo-Inositol
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: INS, MDN, MG, SO4 |
 |
Sulfate-Complex Structure Of A Pyrophosphate-Dependent Kinase In The Ribokinase Family Provides Insight Into The Donor-Binding Mode
Organism: Thermotoga maritima (strain atcc 43589 / msb8 / dsm 3109 / jcm 10099)
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2018-05-16 Classification: TRANSFERASE Ligands: INS, SO4 |
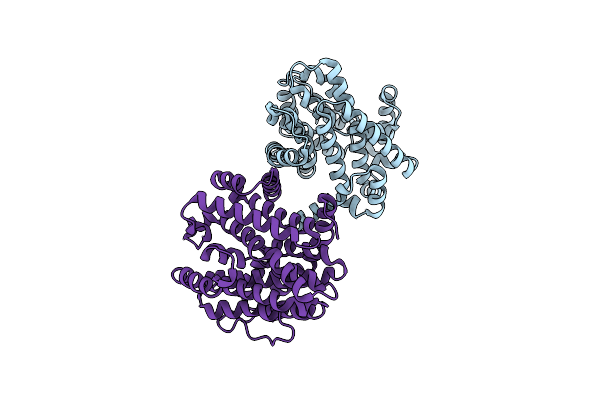 |
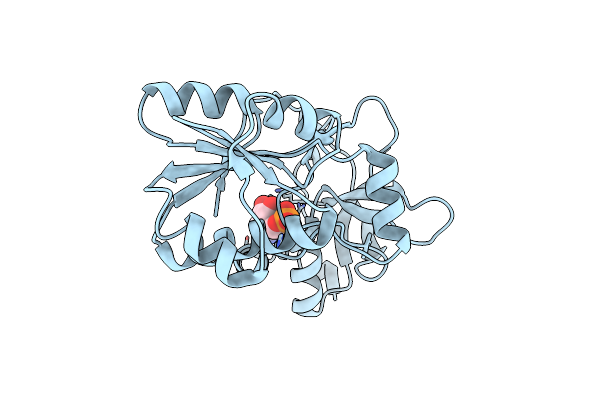
Organism: Bacillus alcalophilus atcc 27647 = cgmcc 1.3604
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-05-09 Classification: LYASE |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP |
 |
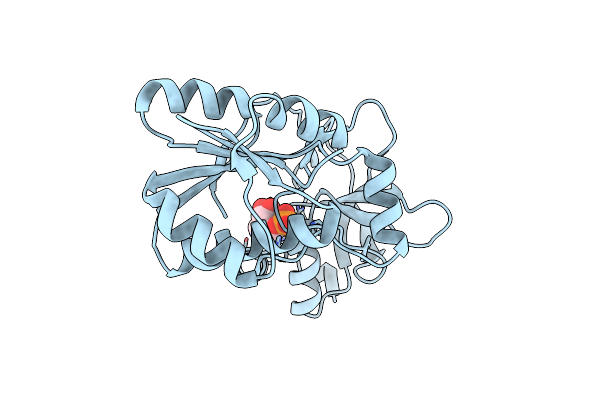
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP, SEP, MG |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP, ADP, 7WF |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP, SEP, MG |
 |
Organism: Thermococcus kodakarensis kod1
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2017-04-12 Classification: TRANSFERASE Ligands: AMP |
 |
Myo-Inositol 3-Kinase Bound With Its Products (Adp And 1D-Myo-Inositol 3-Phosphate)
Organism: Thermococcus kodakaraensis kod1
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2015-06-03 Classification: TRANSFERASE Ligands: LIP, ADP, INS, MG, IOD, EDO |
 |
Organism: Thermococcus kodakaraensis kod1
Method: X-RAY DIFFRACTION Release Date: 2015-06-03 Classification: TRANSFERASE Ligands: INS, ACP, MG, IOD |
 |
Organism: Thermococcus kodakarensis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2015-02-04 Classification: LYASE Ligands: MG, CAP, EDO |
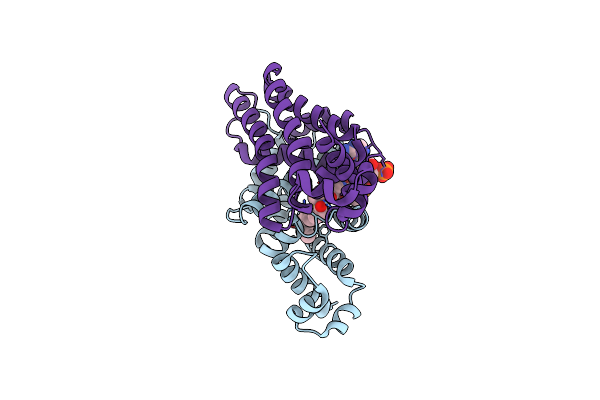 |
Crystal Structure Of Fadr From Bacillus Subtilis, A Transcriptional Regulator Involved In The Regulation Of Fatty Acid Degradation
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: DCC |
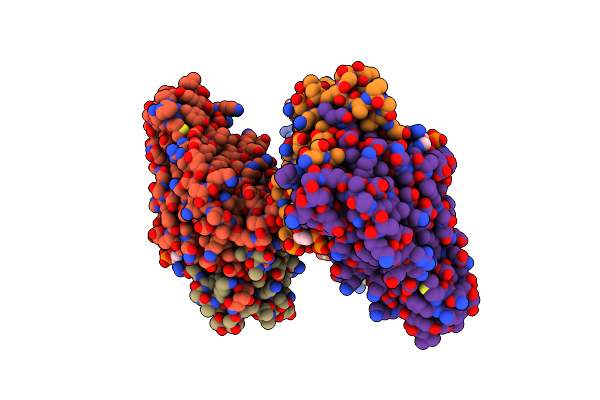 |
Crystal Structure Of A Transcriptional Regulator Fadr From Bacillus Subtilis In Complex With Stearoyl-Coa
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-03-19 Classification: TRANSCRIPTION Ligands: ST9 |
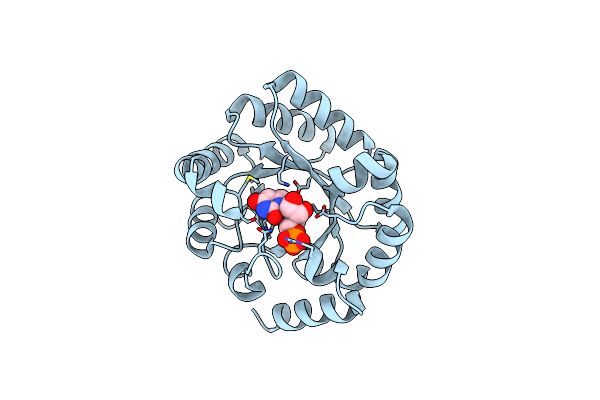 |
Wild-Type Orotidine 5'-Monophosphate Decarboxylase From M. Thermoautotrophicus Complexed With 6-Methyl-Ump
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2013-12-04 Classification: LYASE Ligands: U6M |
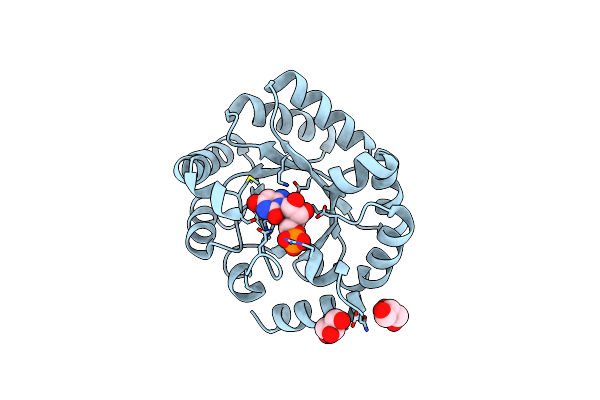 |
Wild-Type Orotidine 5'-Monophosphate Decarboxylase From M. Thermoautotrophicus Complexed With 6-Amino-Ump
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2013-12-04 Classification: LYASE Ligands: NUP, GOL |

