Search Count: 98
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-31 Classification: DNA BINDING PROTEIN/RNA/DNA Ligands: ATP, MG |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-12-03 Classification: DNA BINDING PROTEIN Ligands: ZN |
 |
Organism: Amycolatopsis thermoflava
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: NAP, A1ECB |
 |
Organism: Amycolatopsis thermoflava
Method: X-RAY DIFFRACTION Release Date: 2025-07-23 Classification: OXIDOREDUCTASE Ligands: PGE, PEG, CL, CA, NDP |
 |
Organism: Plasmodium falciparum 3d7
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-05-14 Classification: CELL INVASION |
 |
Organism: Kutzneria sp. 744
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Organism: Kutzneria sp. 744
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, ONH |
 |
Organism: Kutzneria sp. 744
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: HEM, A1ECN |
 |
Organism: Nocardia seriolae
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-11-13 Classification: HYDROLASE Ligands: UMP, PO4, TRS, PEG |
 |
Organism: Thermoflavifilum thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-11-08 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Thermoflavifilum thermophilum
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: DNA BINDING PROTEIN |
 |
Organism: Thermoflavifilum thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Thermoflavifilum thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: DNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Thermoflavifilum thermophilum, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: DNA/RNA/CELL INVASION |
 |
Organism: Canine minute virus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRUS |
 |
Organism: Porcine bocavirus 1 pig/zjd/china/2006
Method: ELECTRON MICROSCOPY Release Date: 2023-08-30 Classification: VIRUS |
 |
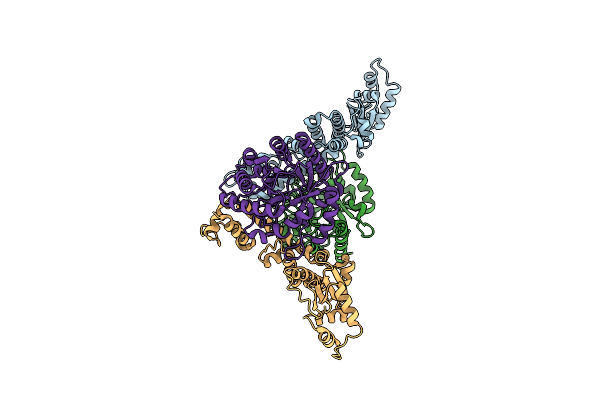 |
Crystal Structure Of A Holoenzyme Fe-Free Tglhi For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2023-08-23 Classification: BIOSYNTHETIC PROTEIN |
 |
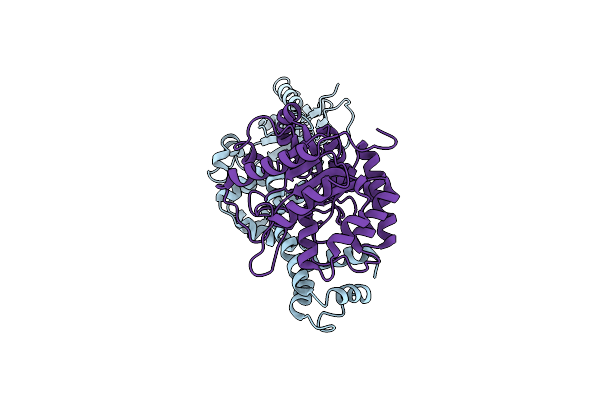
Crystal Structure Of A Holoenzyme Tglhi With Two Fe Irons For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.25 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |
 |
Crystal Structure Of A Holoenzyme Tglhi With Three Fe Ions For Pseudomonas Syringae Peptidyl (S) 2-Mercaptoglycine Biosynthesis
Organism: Pseudomonas syringae pv. maculicola str. es4326
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2023-08-23 Classification: METAL BINDING PROTEIN Ligands: FE |

