Search Count: 46
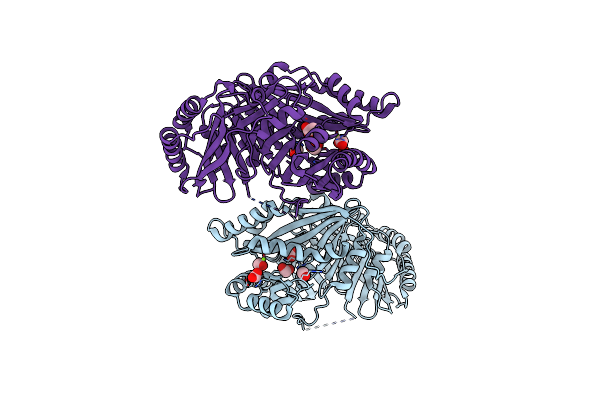 |
Structural Basis Of Chorismate Isomerization By Arabidopsis Isochorismate Synthase Ics1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: ACT, FMT, MG |
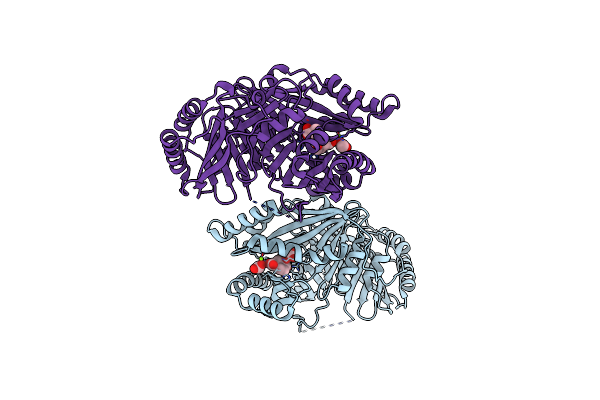 |
Structural Basis Of Chorismate Isomerization By Arabidopsis Isochorismate Synthase Ics1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2024-05-22 Classification: ISOMERASE Ligands: ISJ, MG, FMT |
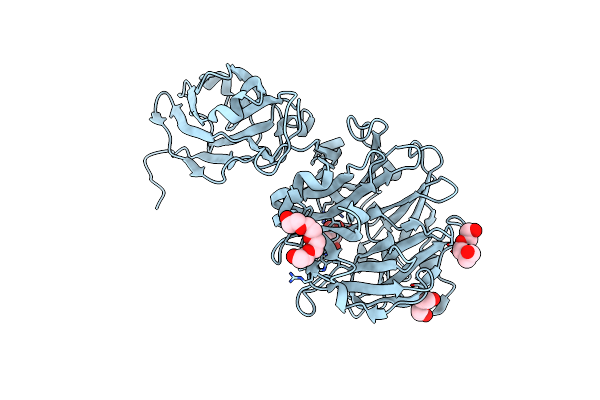 |
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: FLC, PG4, PEG, PGE |
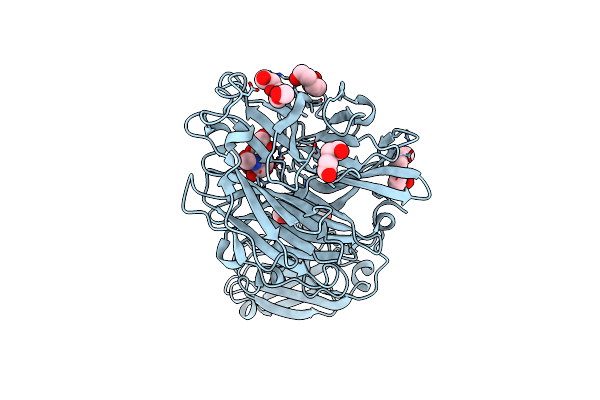 |
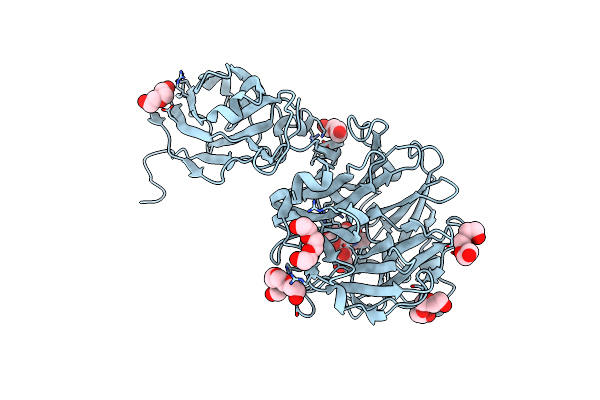
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) Bound To Neu5Ac2En (Dana)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: DAN, PGE, PEG, PG4 |
 |
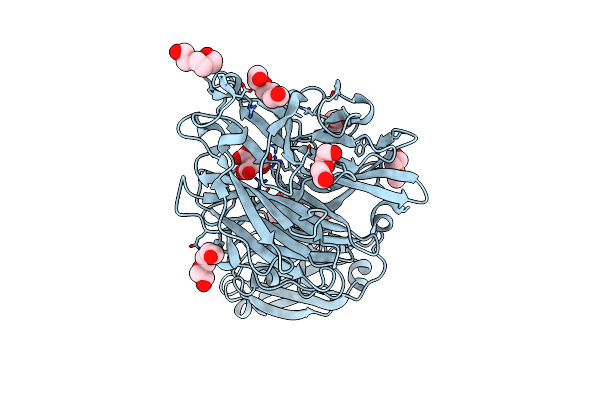
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) Bound To Neu5Ac (Nana)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PGE, PEG, PG4 |
 |
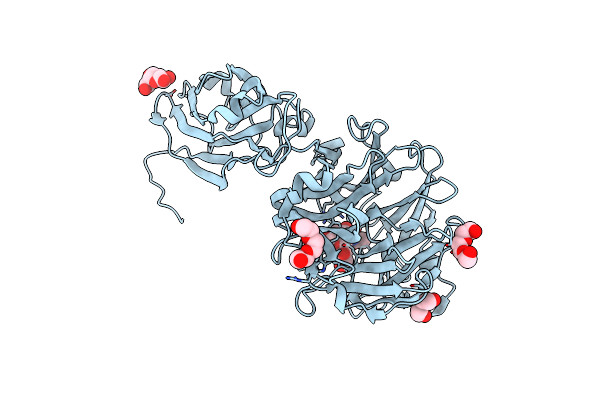
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352)- Fructose Bound In Cbm
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: FLC, PEG, PGE, FUD |
 |
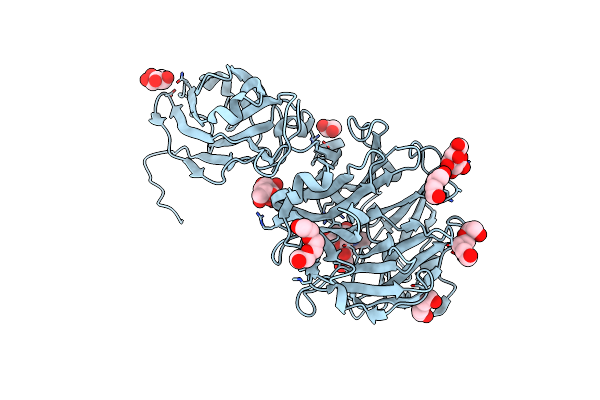
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 3'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, GAL |
 |
Crystal Structure Of Porphyromonas Gingivalis Sialidase (Pg_0352) D219A Mutant Bound To 6'-Sialyllactose (Only Neu5Ac Visible)
Organism: Porphyromonas gingivalis
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2023-10-04 Classification: HYDROLASE Ligands: SIA, PEG, PGE, EDO, GAL |
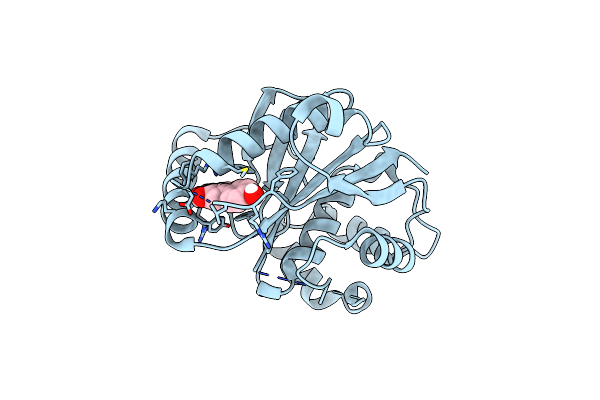 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2023-04-19 Classification: HYDROLASE Ligands: JXY |
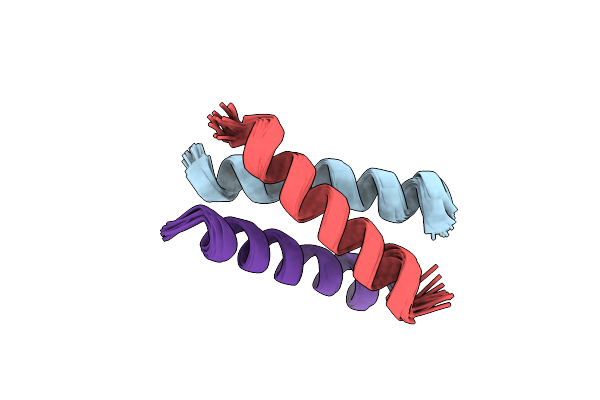 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2021-06-23 Classification: VIRAL PROTEIN |
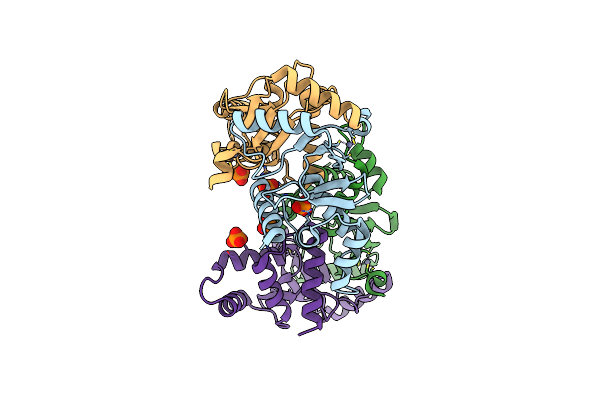 |
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-06-09 Classification: METAL BINDING PROTEIN Ligands: ZN, PO4 |
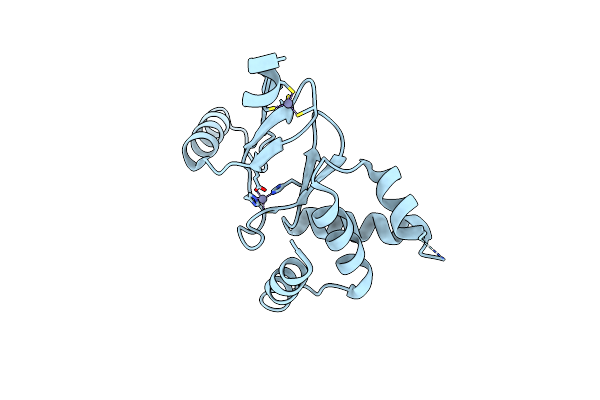 |
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-06-09 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2021-04-28 Classification: MEMBRANE PROTEIN |
 |
Model Of The Hiv-1 Gp41 Membrane-Proximal External Region, Transmembrane Domain And Cytoplasmic Tail
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2021-04-28 Classification: MEMBRANE PROTEIN |
 |
 |
Organism: Agrobacterium tumefaciens a6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-15 Classification: TRANSCRIPTION |
 |
Complex Structure Of A Novel Virulence Regulation Factor Sghr With Its Effector Sucrose
Organism: Agrobacterium tumefaciens a6
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-15 Classification: TRANSCRIPTION |
 |
Organism: Agrobacterium tumefaciens a6, Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2020-07-15 Classification: TRANSCRIPTION |
 |
Structure Of The Hiv-1 Gp41 Transmembrane Domain And Cytoplasmic Tail (Llp2)
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |
 |
Model Of The Hiv-1 Gp41 Membrane-Proximal External Region, Transmembrane Domain And Cytoplasmic Tail (Llp2)
Organism: Human immunodeficiency virus 1
Method: SOLUTION NMR Release Date: 2020-05-13 Classification: MEMBRANE PROTEIN |

