Search Count: 105
 |
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
 |
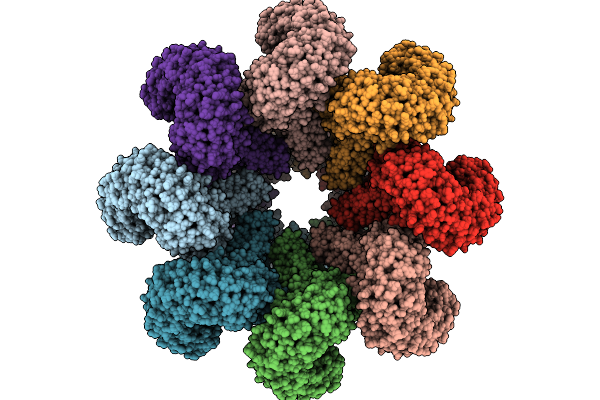
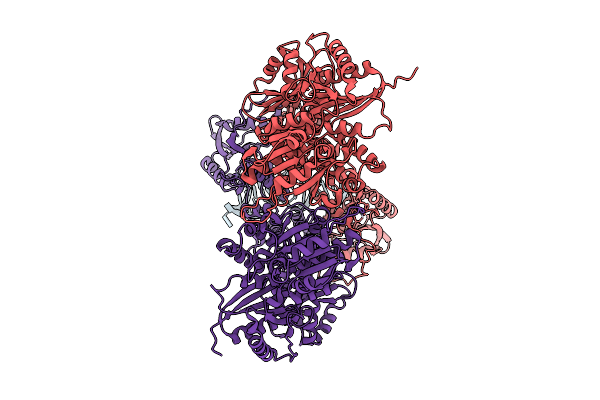
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat (N-To-N Arrangement)
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
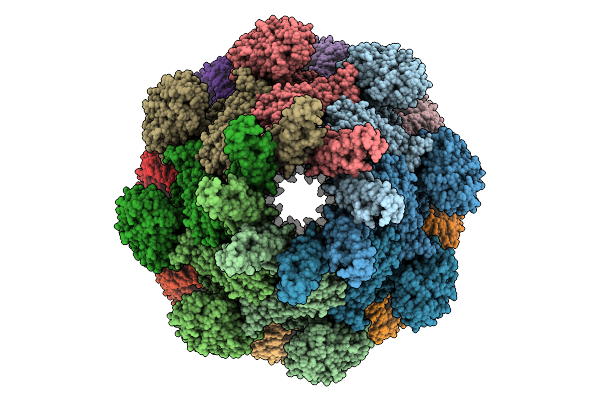 |
Cryo-Em Structure Of An Octameric G10-Resistosome From Wheat In 'Back-To-Back' Arrangement
Organism: Triticum aestivum
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: PLANT PROTEIN Ligands: ATP |
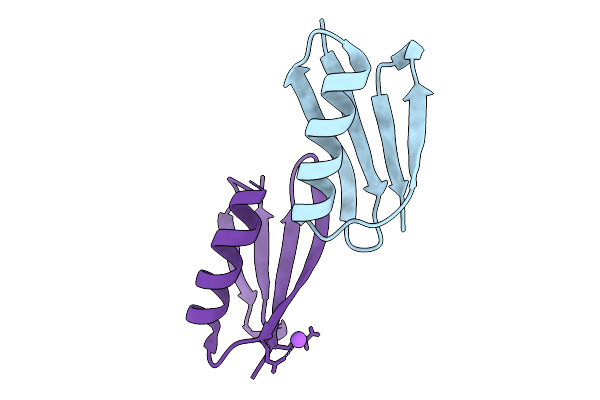 |
X-Ray Structure Of The B1 Domain Of Streptococcal Protein G Triple Mutant T2Q, N8D, And N37D (Gb1-Qdd).
Organism: Streptococcus sp.
Method: X-RAY DIFFRACTION Release Date: 2025-09-03 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN Ligands: ZN |
 |
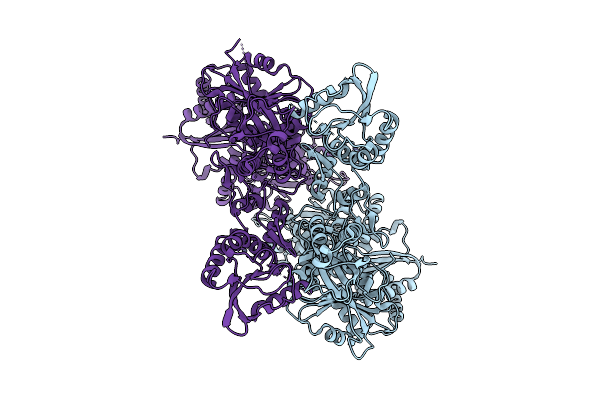
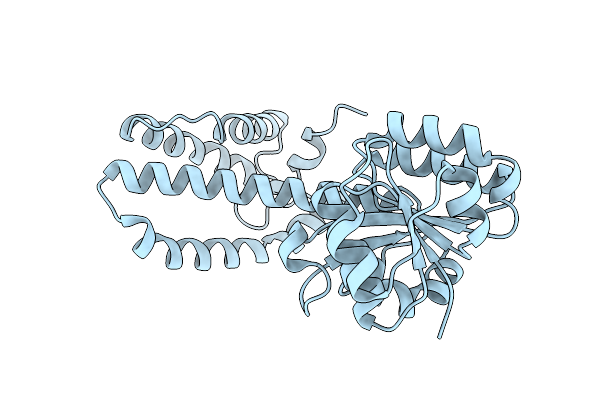
Electronic Microscopy Structure Of Human Schlafen14-E211A Dimer In Complex With Dsrna
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: RNA BINDING PROTEIN/RNA Ligands: ZN |
 |
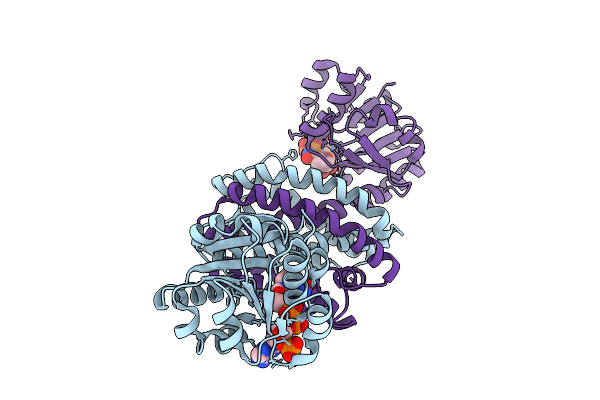
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NAP |
 |
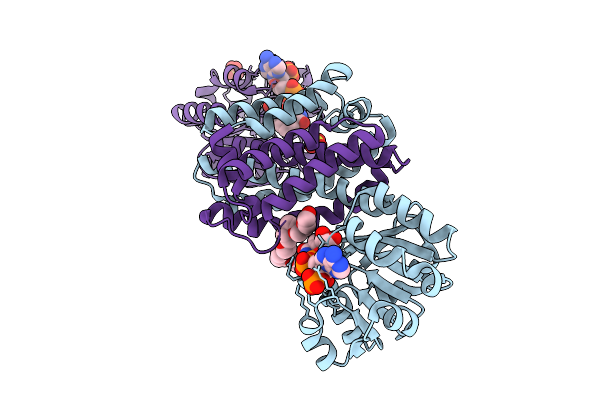
Organism: Micromonospora echinaurantiaca
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, NA |
 |
Organism: Kutzneria albida dsm 43870
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: OXIDOREDUCTASE Ligands: NDP, SO4, PE8, NA |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: VIRAL PROTEIN/IMMUNE SYSTEM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: ALLERGEN/IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-11-27 Classification: SIGNALING PROTEIN Ligands: A1LXY |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT |
 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
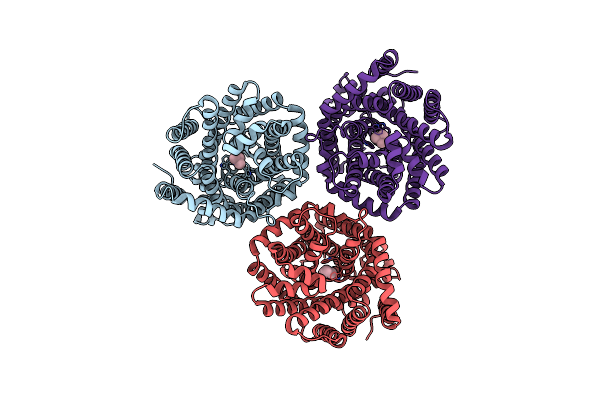 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
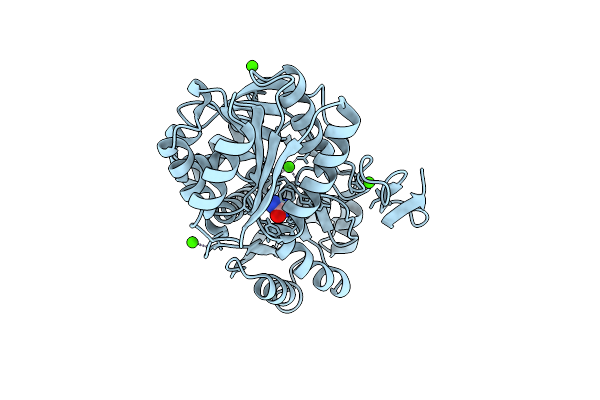 |
Crystal Structure Of Urta From Prochlorococcus Marinus Str. Mit 9313 In Complex With Urea And Calcium
Organism: Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: URE, CA |
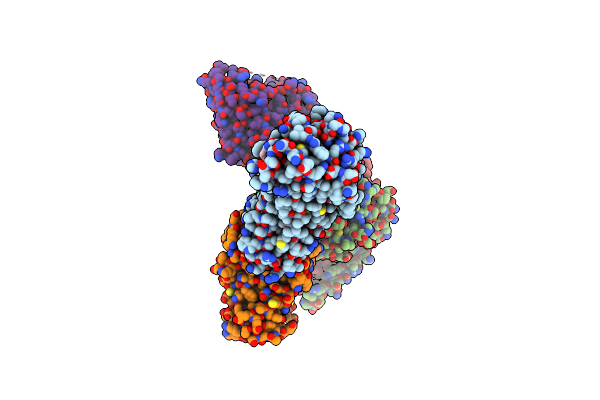 |
Cryo-Em Structure Of Compound 9N Bound Ketone Body Receptor Hcar2-Gi Signaling Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: IX8, CLR |
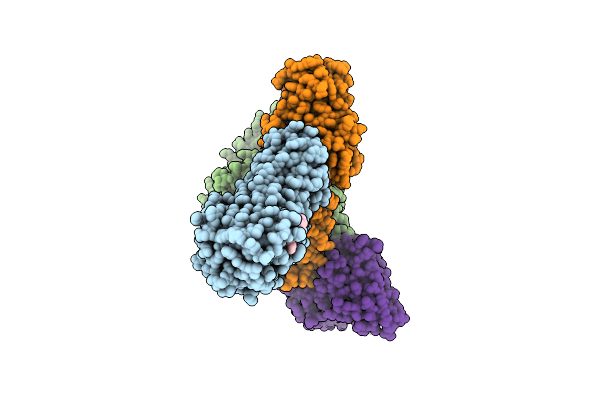 |
Cryo-Em Structure Of Compound 9N And Niacin Bound Ketone Body Receptor Hcar2-Gi Signaling Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: NIO, CLR, IX8 |
 |
Cryo-Em Structure Of Niacin Bound Ketone Body Receptor Hcar2-Gi Signaling Complex
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-09-06 Classification: MEMBRANE PROTEIN Ligands: NIO, CLR |

