Search Count: 50
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/RNA Ligands: MG |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN Ligands: MG, ZN |
 |
Organism: Flagellimonas taeanensis
Method: ELECTRON MICROSCOPY Release Date: 2025-12-24 Classification: RNA BINDING PROTEIN/DNA/RNA Ligands: MG, ZN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
 |
Organism: Streptomyces
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: BIOSYNTHETIC PROTEIN Ligands: F42 |
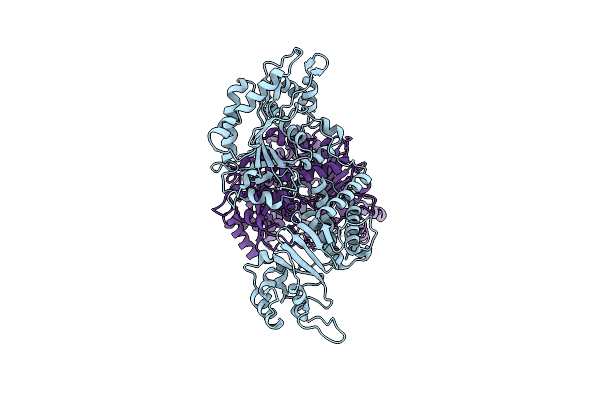 |
Cryo-Electron Microscopy (Cryo-Em) Structure Of The Hachiman Defense System From Escherichia Coli
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-03-26 Classification: DNA BINDING PROTEIN |
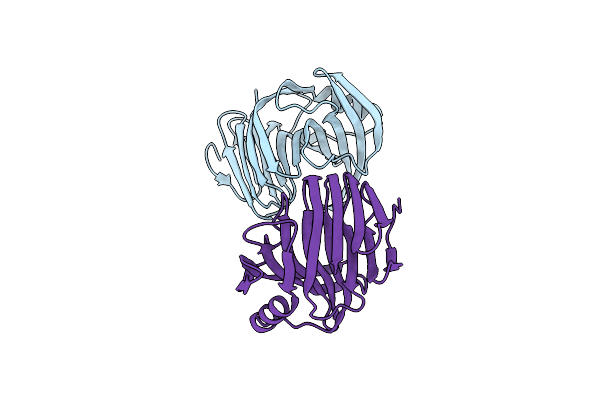 |
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-11-27 Classification: HYDROLASE |
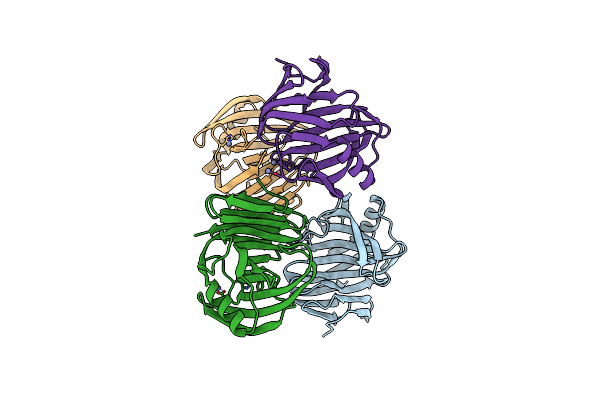 |
Organism: Aspergillus terreus (strain nih 2624 / fgsc a1156)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: ZN |
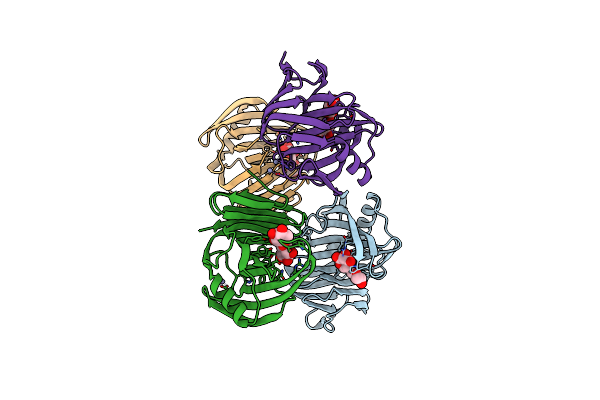 |
Organism: Aspergillus terreus nih2624
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: ZN, XYP |
 |
Organism: Aspergillus terreus nih2624
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2024-11-27 Classification: HYDROLASE Ligands: ZN, XYP |
 |
Organism: Ovis aries
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: TRANSPORT PROTEIN |
 |
Organism: Xenopus laevis
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: TRANSPORT PROTEIN |
 |
The Sigma-1 Receptor From Xenopus Laevis In Complex With Progesterone By Co-Crystallization
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-07-17 Classification: MEMBRANE PROTEIN Ligands: STR |
 |
The Sigma-1 Receptor From Xenopus Laevis In Complex With Progesterone By Soaking
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2024-07-17 Classification: MEMBRANE PROTEIN Ligands: STR |
 |
Crystal Structure Of The Sigma-1 Receptor From Xenopus Laevis In The Absence Of Known Ligands
Organism: Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:2.17 Å Release Date: 2024-07-17 Classification: MEMBRANE PROTEIN |

