Search Count: 91
 |
A Lid Blocking Mechanism Of A Cone Snail Toxin Revealed At The Atomic Level
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-07-14 Classification: TOXIN |
 |
Determinants Of Repressor/Operator Recognition From The Structure Of The Trp Operator Binding Site
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-10-14 Classification: DNA |
 |
Pore-Modulating Toxins Exploit Inherent Slow Inactivation To Block K+ Channels
Organism: Conus striatus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-08-21 Classification: TOXIN Ligands: SO4 |
 |
Pore-Modulating Toxins Exploit Inherent Slow Inactivation To Block K+ Channels
Organism: Conus striatus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2019-08-21 Classification: TOXIN Ligands: PO4, NO3, EDO |
 |
Organism: Clostridium clariflavum
Method: X-RAY DIFFRACTION Resolution:1.13 Å Release Date: 2019-06-12 Classification: SUGAR BINDING PROTEIN Ligands: EDO, CA |
 |
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2019-06-12 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
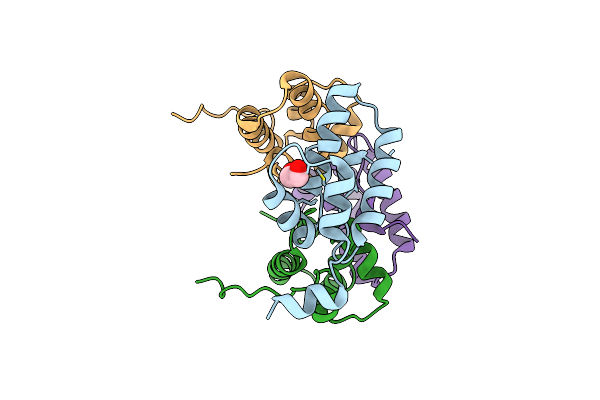
Crystal Structure Of The Human Mitochondrial Chaperonin Symmetrical 'Football' Complex
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2015-04-29 Classification: CHAPERONE Ligands: ADP, MG |
 |
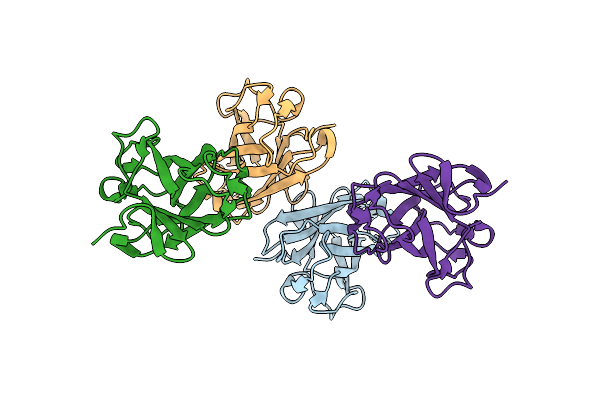
Structure Of A Novel Autonomous Cohesin Protein From Ruminococcus Flavefaciens
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2013-12-18 Classification: STRUCTURAL PROTEIN Ligands: CL |
 |
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2013-10-16 Classification: VIRAL PROTEIN Ligands: EDO |
 |
Crystal Structure Of Carbohydrate-Binding Module Cbm3B Mutant (Y56S) From The Cellulosomal Cellobiohydrolase 9A From Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-10-09 Classification: HYDROLASE |
 |
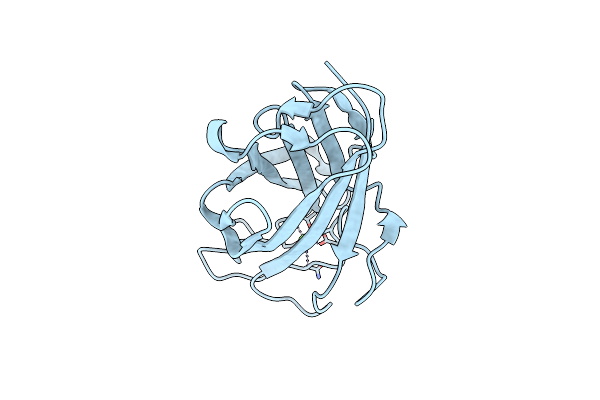
Family 3B Carbohydrate-Binding Module From The Biomass Sensoring System Of Clostridium Clariflavum
Organism: Clostridium clariflavum
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2013-09-18 Classification: SUGAR BINDING PROTEIN Ligands: CL, CA |
 |
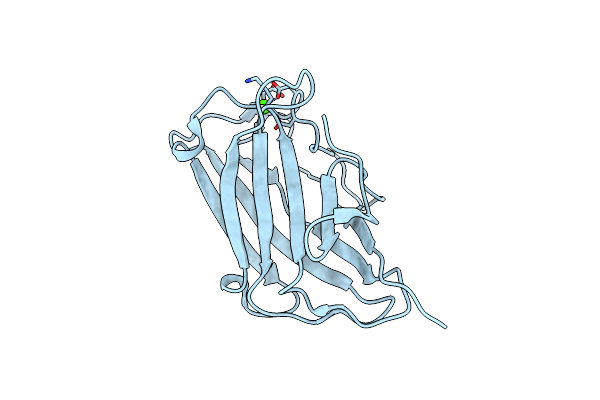
Biomass Sensing Modules From Putative Rsgi-Like Proteins Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2013-09-11 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Biomass Sensoring Modules From Putative Rsgi-Like Proteins Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.17 Å Release Date: 2013-09-11 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CA |
 |
Biomass Sensoring Module From Putative Rsgi2 Protein Of Clostridium Thermocellum Resemble Family 3 Carbohydrate-Binding Module Of Cellulosome
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.18 Å Release Date: 2013-09-11 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CA, ZN |
 |
Cbm3A-L Domain With Flanking Linkers From Scaffoldin Cipa Of Cellulosome Of Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2013-07-17 Classification: CELLULOSE BINDING PROTEIN Ligands: CA, GOL, SO4, CL, 1MZ |
 |
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-04-24 Classification: STRUCTURAL PROTEIN Ligands: CA, SO4, CL |
 |
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2013-04-24 Classification: STRUCTURAL PROTEIN Ligands: CA, SO4 |
 |
Crystal Structure Of An Uncommon Cellulosome-Related Protein Module From Ruminococcus Flavefaciens That Resembles Papain-Like Cysteine Peptidases
Organism: Ruminococcus flavefaciens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2013-03-20 Classification: HYDROLASE Ligands: EDO |
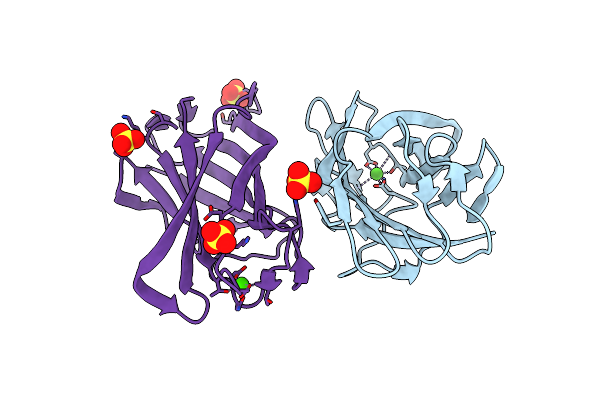 |
High Resolution Structure For Family 3A Carbohydrate Binding Module From The Cipa Scaffolding Of Clostridium Thermocellum
Organism: Clostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2012-09-12 Classification: SUGAR BINDING PROTEIN Ligands: CA, SO4 |
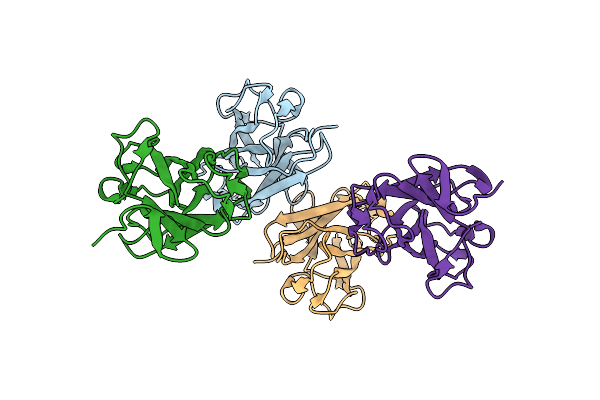 |
Carbohydrate-Binding Module Cbm3B From The Cellulosomal Cellobiohydrolase 9A From Clostridium Thermocellum
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2012-04-25 Classification: HYDROLASE |

