Search Count: 53
 |
Sialic Acid Bound Form Of Tripartite Atp-Independent Periplasmic (Trap) Transporter From Fusobacterium Nucleatum.
Organism: Fusobacterium nucleatum, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Apo Form Of Tripartite Atp-Independent Periplasmic (Trap) Transporter From Fusobacterium Nucleatum.
Organism: Fusobacterium nucleatum, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
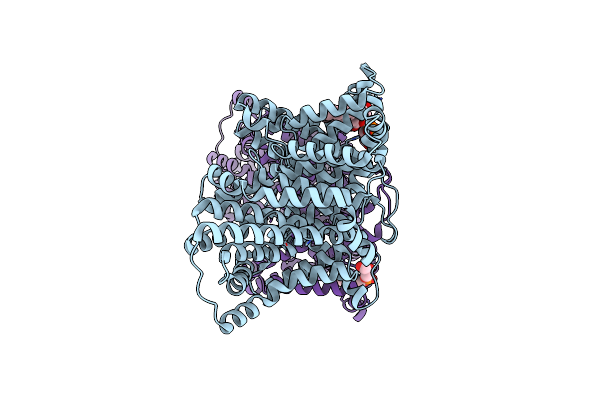 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Parallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: PGT, NA |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Haemophilus Influenzae (Antiparallel Dimer)
Organism: Haemophilus influenzae rd kw20
Method: ELECTRON MICROSCOPY Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In A Nanodisc
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-03-15 Classification: TRANSPORT PROTEIN Ligands: OCT, PTY, TWT, D10, NA, TRD, HEX |
 |
Cryo-Em Structure Of The Tripartite Atp-Independent Periplasmic (Trap) Transporter Siaqm From Photobacterium Profundum In Amphipol
Organism: Photobacterium profundum ss9, Helicobacter pylori, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2022-12-21 Classification: TRANSPORT PROTEIN |
 |
Structure Of The Sialic Acid Bound Tripartite Atp-Independent Periplasmic (Trap) Periplasmic Component Siap From Photobacterium Profundum
Organism: Photobacterium profundum
Method: X-RAY DIFFRACTION Resolution:1.04 Å Release Date: 2022-12-14 Classification: TRANSPORT PROTEIN Ligands: SLB, SO4 |
 |
N-Acetylmannosamine-6-Phosphate 2-Epimerase From Staphylococcus Aureus (Strain Mrsa Usa300) With 5-Deoxy Substrate Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-04-20 Classification: ISOMERASE Ligands: ZM1, CIT, CL |
 |
N-Acetylmannosamine-6-Phosphate 2-Epimerase E180A From Staphylococcus Aureus (Strain Mrsa Usa300)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2022-04-13 Classification: ISOMERASE Ligands: CIT, CL, EDO |
 |
N-Acetylmannosamine-6-Phosphate 2-Epimerase From Staphylococcus Aureus (Strain Mrsa Usa300) With Substrate And Product Bound
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-04-13 Classification: ISOMERASE Ligands: CL, LRY, RFW |
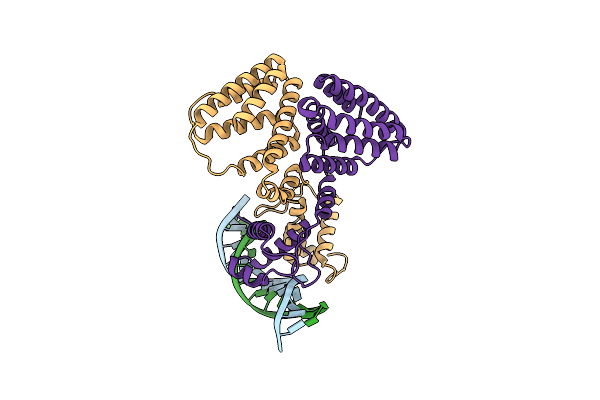 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: GENE REGULATION |
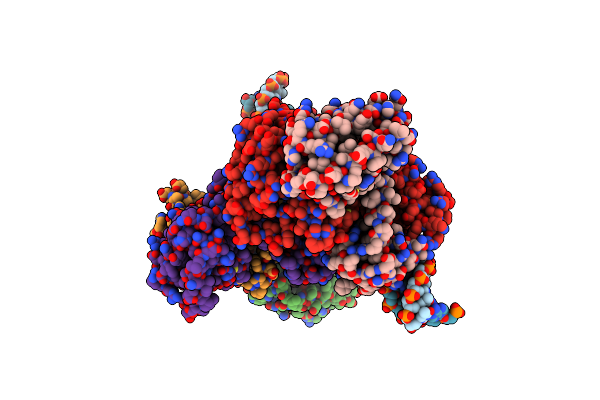 |
Coordinates Of Nanr Dimer Fitted In Hexameric Nanr-Dna Hetero-Complex Cryo-Em Map
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-10 Classification: GENE REGULATION |
 |
N-Acetylmannosamine-6-Phosphate 2-Epimerase From Staphylococcus Aureus (Strain Mrsa Usa300)
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2021-02-24 Classification: ISOMERASE Ligands: CIT, CL |
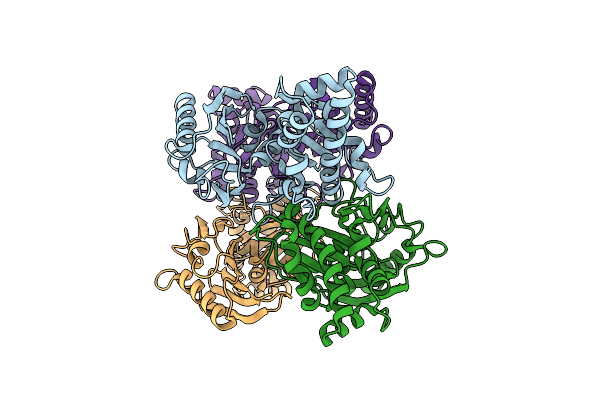 |
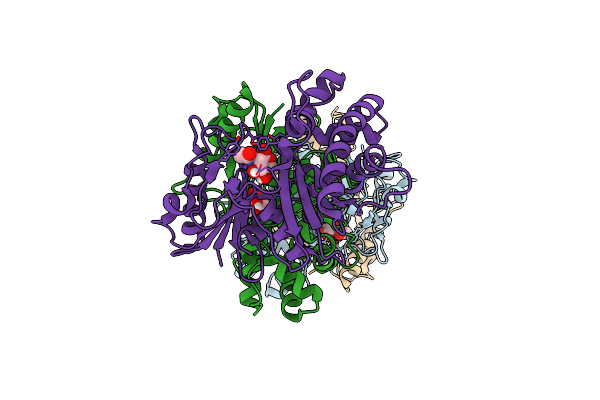
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2020-01-22 Classification: TRANSFERASE |
 |
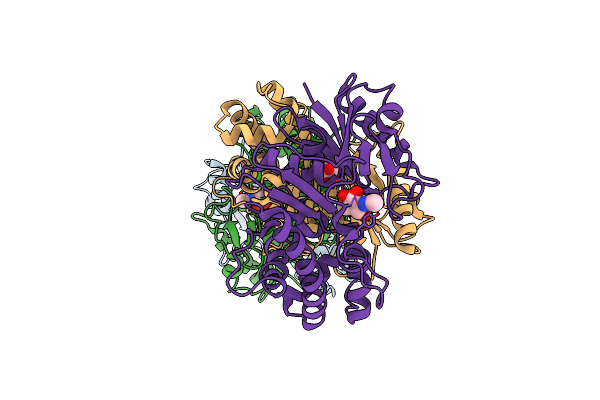
N-Acetylmannosamine Kinase With N-Acetylmannosamine From Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-22 Classification: TRANSFERASE Ligands: BM3 |
 |
Metal Rok Rebel: Characterisation Of N-Acetylmannosamine Kinase From The Pathogen Staphylococcus Aureus
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-22 Classification: TRANSFERASE Ligands: NAG |
 |
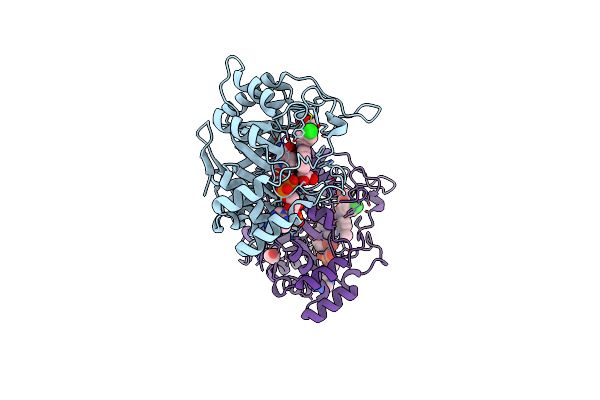
Organism: Proteus mirabilis (strain hi4320)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2019-06-26 Classification: LYASE Ligands: SO4 |
 |
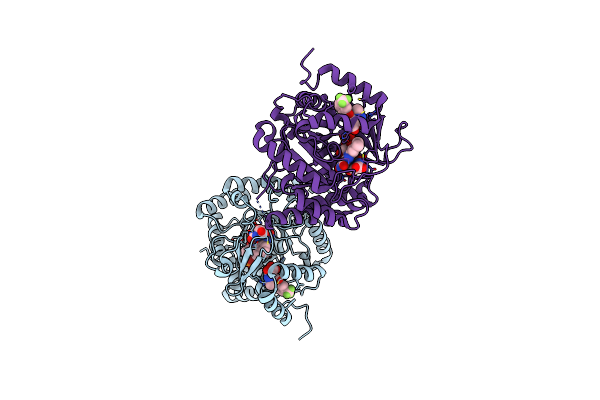
Crystal Structure Of Aldo-Keto Reductase 1C3 (Akr1C3) Complexed With Inhibitor.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2019-05-08 Classification: OXIDOREDUCTASE Ligands: NAP, FFW, EDO |
 |
Plasmodium Falciparum Dihydroorotate Dehydrogenase (Dhodh) Co-Crystallized With 3-Hydroxy-1-Methyl-5-((3-(Trifluoromethyl)Phenoxy)Methyl)-1H-Pyrazole-4-Carboxylic Acid
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: FMN, ORO, E2N |
 |
Plasmodium Falciparum Dihydroorotate Dehydrogenase (Dhodh) Co-Crystallized With N-(2,2-Diphenylethyl)-4-Hydroxy-1,2,5-Thiadiazole-3-Carboxamide
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2018-12-19 Classification: OXIDOREDUCTASE Ligands: FMN, ORO, DZB |

