Search Count: 18
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN Ligands: MG, ANP |
 |
Organism: Escherichia coli (strain k12)
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN Ligands: MG, ADP, VO4 |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN Ligands: ADP |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.30 Å Release Date: 2022-01-12 Classification: DNA BINDING PROTEIN Ligands: MG, ATP, ADP |
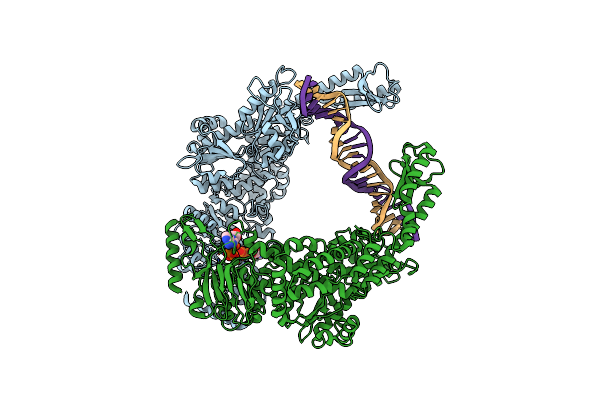 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:4.40 Å Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN Ligands: ATP |
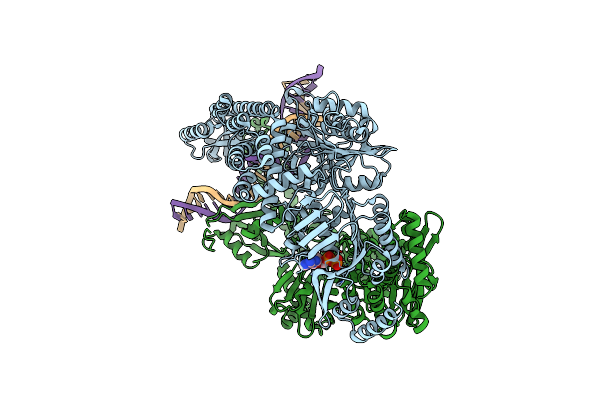 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN Ligands: ADP |
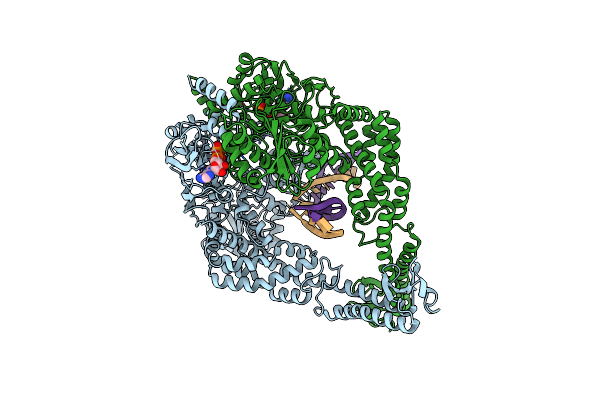 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN Ligands: ADP |
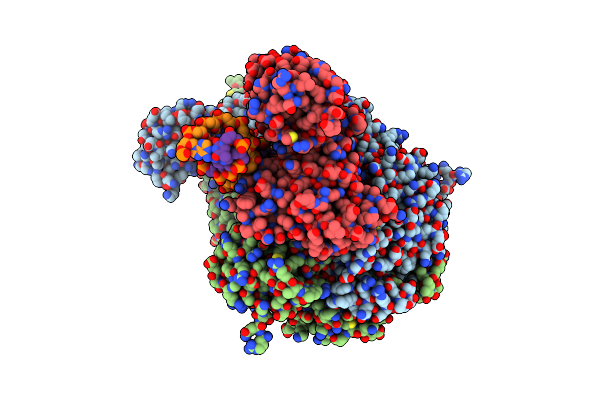 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN Ligands: ANP |
 |
Organism: Escherichia coli (strain k12), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2021-03-31 Classification: DNA BINDING PROTEIN Ligands: ANP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-04-24 Classification: RNA BINDING PROTEIN |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:4.71 Å Release Date: 2015-07-22 Classification: DNA BINDING PROTEIN Ligands: ANP |
 |
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2015-07-22 Classification: HYDROLASE Ligands: ANP |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:7.60 Å Release Date: 2015-07-22 Classification: DNA BINDING PROTEIN Ligands: ANP |
 |
Crystal Structure Of Full-Length E.Coli Dna Mismatch Repair Protein Muts D835R Mutant In Complex With Gt Mismatched Dna
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-07-17 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2010-12-08 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-07-21 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-07-21 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2010-07-21 Classification: HYDROLASE Ligands: ZN |

