Search Count: 38
 |
Organism: Corticium sp. (in: fungi)
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2022-07-13 Classification: SUGAR BINDING PROTEIN Ligands: MAN, CA |
 |
Organism: Corticium sp. (in: fungi)
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Corticium sp. (in: fungi)
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: MG |
 |
Organism: Corticium sp. (in: fungi)
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: FUC, CA |
 |
Organism: Corticium sp. (in: fungi)
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2022-07-06 Classification: SUGAR BINDING PROTEIN Ligands: CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2019-10-09 Classification: TRANSPORT PROTEIN Ligands: GTP, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.48 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: MG, GCP, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP, MG, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP, MG, CL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2016-09-21 Classification: HYDROLASE Ligands: GDP, MG |
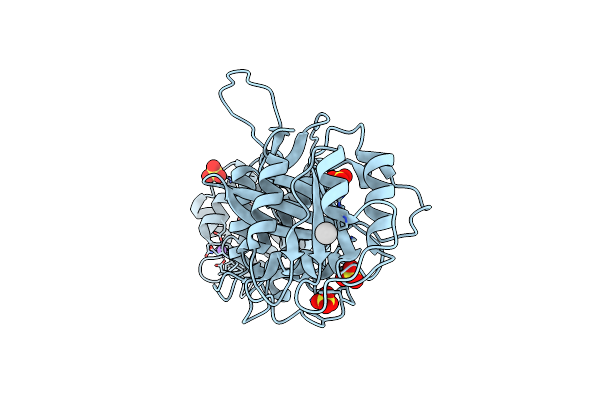 |
Crystal Structure Of Drosophila Miro Ef Hand And Cgtpase Domains In The Apo State (Apo-Miros)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.82 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: UNX, SO4, NA, HSE |
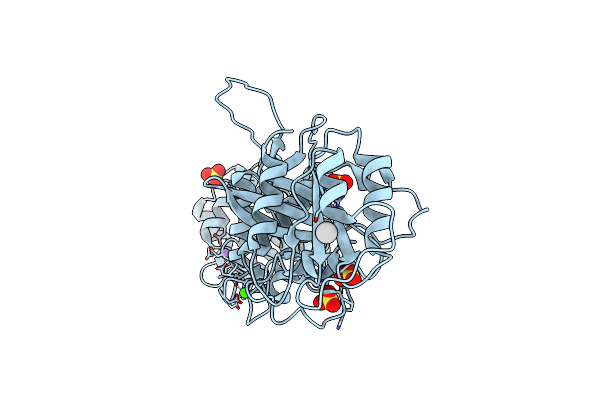 |
Crystal Structure Of Drosophila Miro Ef Hand And Cgtpase Domains Bound To One Calcium Ion (Ca-Miros)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: HSE, UNX, SO4, NA, CA |
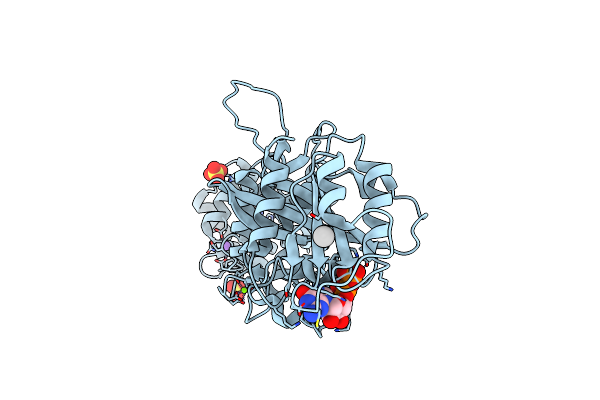 |
Crystal Structure Of Drosophila Miro Ef Hand And Cgtpase Domains Bound To One Magnesium Ion And Mg:Gdp (Mggdp-Miros)
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2013-10-09 Classification: HYDROLASE Ligands: UNX, SO4, NA, GDP, MG, HSE |
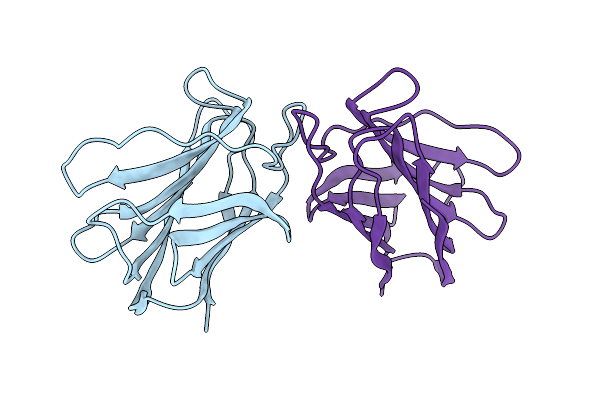 |
Organism: Cinachyrella sp.
Method: X-RAY DIFFRACTION Resolution:2.98 Å Release Date: 2012-08-29 Classification: CARBOHYDRATE-BINDING PROTEIN Ligands: CL |
 |
Organism: Cinachyrella
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2012-08-29 Classification: SUGAR BINDING PROTEIN Ligands: CL |
 |
Organism: Cinachyrella
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2012-08-29 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Thermus aquaticus
Method: X-RAY DIFFRACTION Resolution:1.24 Å Release Date: 2007-02-13 Classification: SIGNALING PROTEIN Ligands: GDP, EDO, DIO |

