Search Count: 21
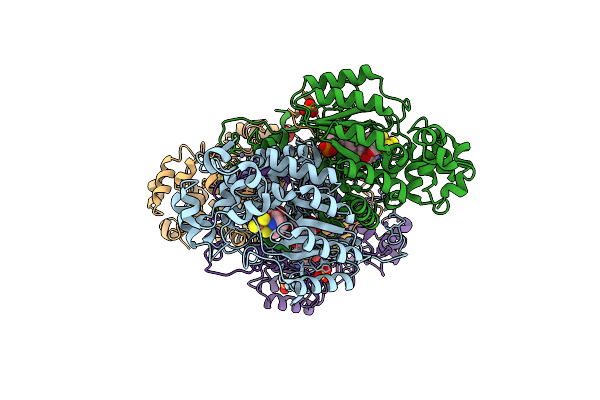 |
2.1 Angstrom X-Ray Crystal Structure Of Lysine-2,3-Aminomutase From Clostridium Subterminale Sb4, With Michaelis Analog (L-Alpha-Lysine External Aldimine Form Of Pyridoxal-5'-Phosphate).
Organism: Clostridium subterminale
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2005-10-04 Classification: ISOMERASE Ligands: ZN, SO4, SAM, LYS, PLP, SF4 |
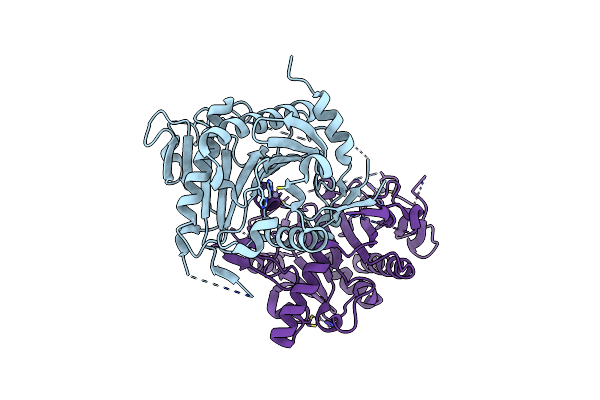 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-06-14 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN |
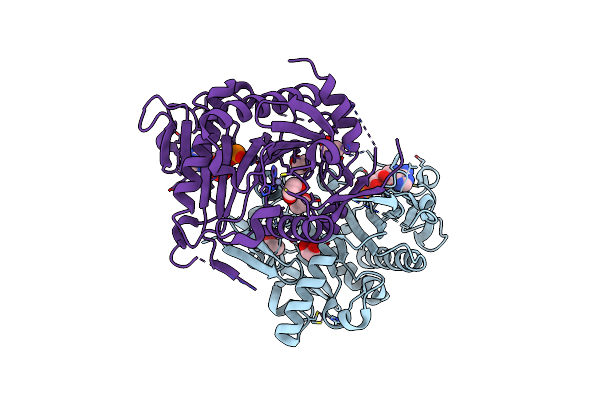 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2005-04-19 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION Ligands: ZN, AMP, EDO |
 |
Structure And A Proposed Mechanism For Ornithine Cyclodeaminase From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2004-11-09 Classification: LYASE |
 |
Crystal Structure Analysis Of Ornithine Cyclodeaminase Complexed With Nad And Ornithine To 1.6 Angstroms
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2004-11-09 Classification: LYASE |
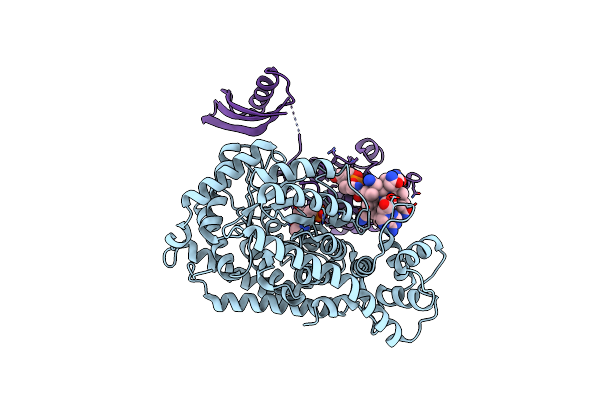 |
Crystal Structure Of Lysine 5,6-Aminomutase In Complex With Plp, Cobalamin, And 5'-Deoxyadenosine
Organism: Clostridium sticklandii
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-11-09 Classification: ISOMERASE Ligands: B12, PLP, 5AD |
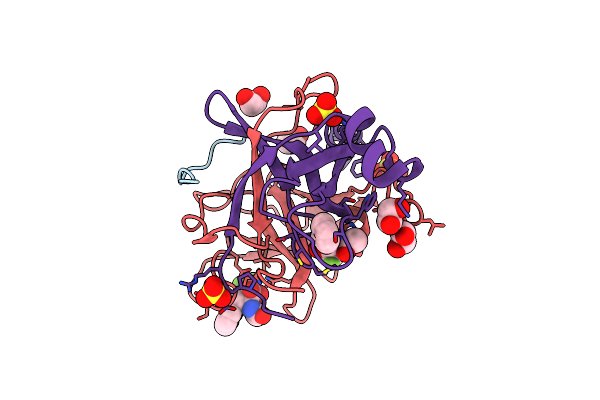 |
Crystal Structure Of Gamma Chymotrypsin With N-Acetyl-Phenylalanine Trifluoromethyl Ketone Bound At The Active Site
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2000-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, APL, EDO, APF |
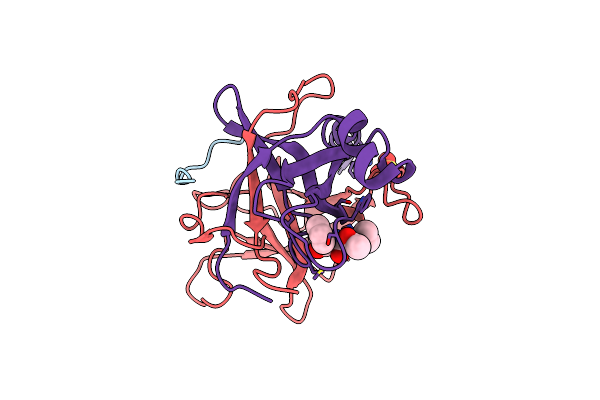 |
Crystal Structure Of Gamma Chymotrypsin With N-Acetyl-Leucil-Phenylalanine Aldehyde Bound At The Active Site
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2000-09-20 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: SO4, FAF |
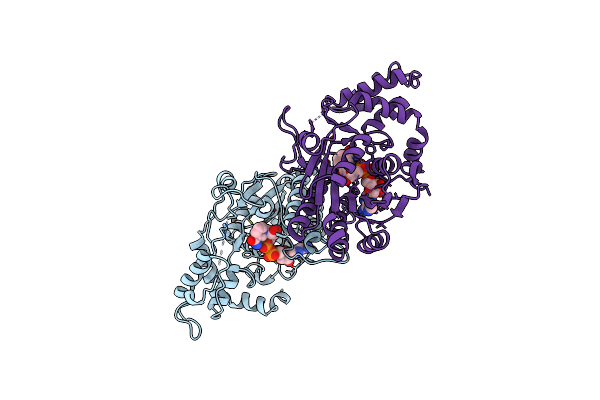 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-10-14 Classification: LYASE Ligands: NAD |
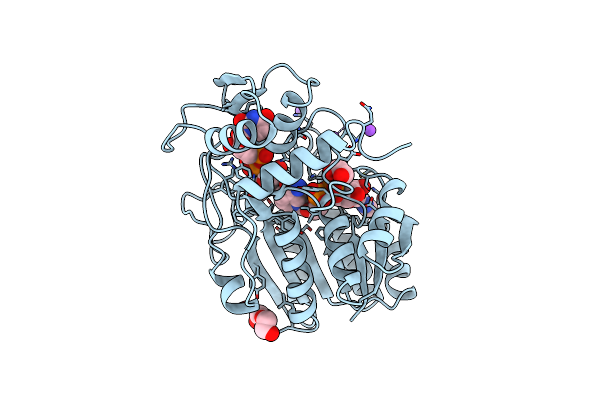 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 1998-03-18 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-03-18 Classification: ISOMERASE Ligands: NAD, UPP, NA, EDO |
 |
Structure Of Udp-Galactose-4-Epimerase Complexed With Udp-4-Deoxy-4-Fluoro-Alpha-D-Galactose
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1998-01-14 Classification: ISOMERASE |
 |
Structure Of Udp-Galactose-4-Epimerase Complexed With Udp-4-Deoxy-4-Fluoro-Alpha-D-Glucose
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1998-01-14 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 1998-01-14 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-11-12 Classification: NUCLEOTIDYLTRANSFERASE Ligands: GDU, ZN, FE, K |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-11-12 Classification: NUCLEOTIDYLTRANSFERASE Ligands: ZN, FE, K, UPG |
 |
The Structure Of Nucleotidylated Galactose-1-Phosphate Uridylyltransferase From Escherichia Coli At 1.86 Angstroms Resolution
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 1997-10-22 Classification: NUCLEOTIDYLTRANSFERASE Ligands: ZN, FE, U5P |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1997-02-12 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 1996-12-23 Classification: ISOMERASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1996-12-23 Classification: ISOMERASE |

