Search Count: 77
 |
Crystal Structure Of The Vim-2 Acquired Metallo-Beta-Lactamase In Complex With Jmv-4690 (Cpd 31)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-09-30 Classification: HYDROLASE Ligands: ZN, PJB, EDO, ACT, DMS |
 |
Fez-1 Metallo-Beta-Lactamase From Legionella Gormanii Modelled With Unknown Ligand
Organism: Fluoribacter gormanii
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: ZN, SO4, GOL, UNL |
 |
Organism: Fluoribacter gormanii
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-06-20 Classification: HYDROLASE Ligands: ZN, SO4, GOL, UNX |
 |
1,2,4-Triazole-3-Thione Compounds As Inhibitors Of L1, Di-Zinc Metallo-Beta-Lactamases.
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2017-01-11 Classification: HYDROLASE Ligands: ZN, L3B, SO4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2016-02-03 Classification: HYDROLASE Ligands: ZN |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-02-03 Classification: HYDROLASE Ligands: ZN, ACT |
 |
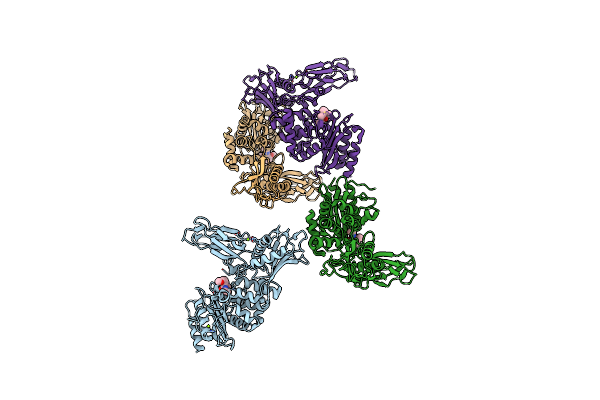
Crystal Structure Of Fox-4 Cephamycinase Mutant Y150F Complexed With Cefoxitin
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2016-02-03 Classification: HYDROLASE Ligands: 1S7, ZN, NA |
 |
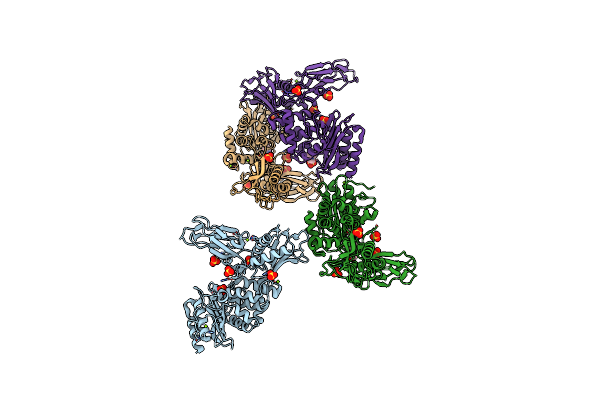
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp.r39
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2012-02-29 Classification: HYDROLASE Ligands: B07, SO4, MG |
 |
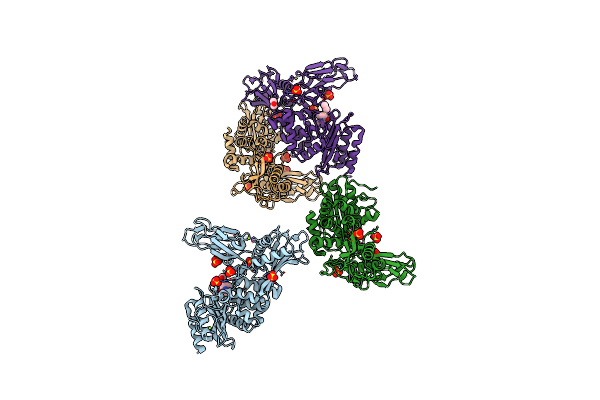
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2012-02-29 Classification: HYDROLASE Ligands: SO4, MG, FP5, ACN, 33D, MES |
 |
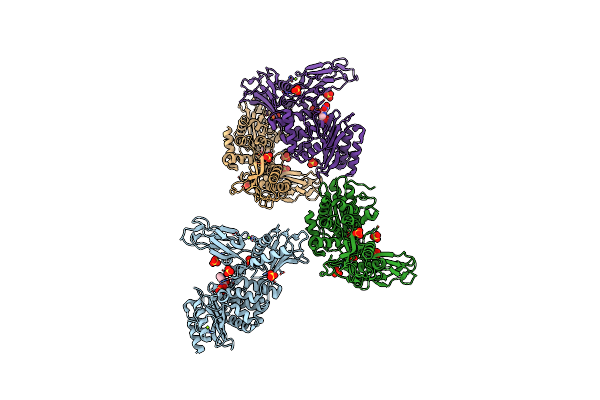
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2011-07-27 Classification: HYDROLASE Ligands: BH6, SO4, MG |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Release Date: 2011-07-27 Classification: HYDROLASE Ligands: FP5, SO4, MG, ACN |
 |
Unexpected Tricovalent Binding Mode Of Boronic Acids Within The Active Site Of A Penicillin Binding Protein
Organism: Actinomadura sp. r39
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-07-27 Classification: HYDROLASE Ligands: ZA3, SO4, MG |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.79 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: ZZ7, SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: SO4, GOL, EDO |
 |
Crystal Structure Of The Mono-Zinc Metallo-Beta-Lactamase Vim-4 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, FLC, CIT |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, IFS, SO4, GOL |
 |
Organism: Aeromonas hydrophila
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2010-06-23 Classification: HYDROLASE Ligands: ZN, GOL, SO4, SDF |
 |
Crystal Structure Of The Oxa-10 V117T Mutant At Ph 6.5 Inhibited By A Chloride Ion
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: GOL, CL, CIT, SO4 |
 |
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: SO4, GOL |
 |
Crystal Structure Of The Di-Zinc Metallo-Beta-Lactamase Vim-4 From Pseudomonas Aeruginosa
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-05-19 Classification: HYDROLASE Ligands: ZN, FLC |

