Search Count: 19
 |
Organism: Galaxea fascicularis
Method: X-RAY DIFFRACTION Resolution:1.39 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN Ligands: CL |
 |
Organism: Acropora millepora
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN Ligands: CL, BR |
 |
Organism: Echinopora forskaliana
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN |
 |
Organism: Stylophora pistillata
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2022-04-20 Classification: FLUORESCENT PROTEIN |
 |
An Unexpected Vestigial Protein Complex Reveals The Evolutionary Origins Of An S-Triazine Catabolic Enzyme.
Organism: Pseudomonas sp. (strain adp)
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: MG |
 |
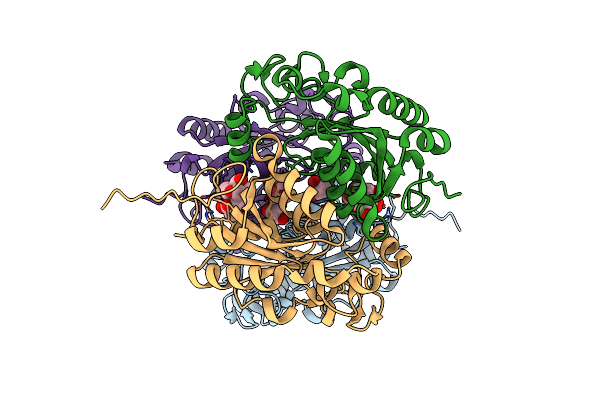
An Unexpected Vestigial Protein Complex Reveals The Evolutionary Origins Of An S-Triazine Catabolic Enzyme. Inhibitor Bound Complex.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-03-21 Classification: HYDROLASE Ligands: CA |
 |
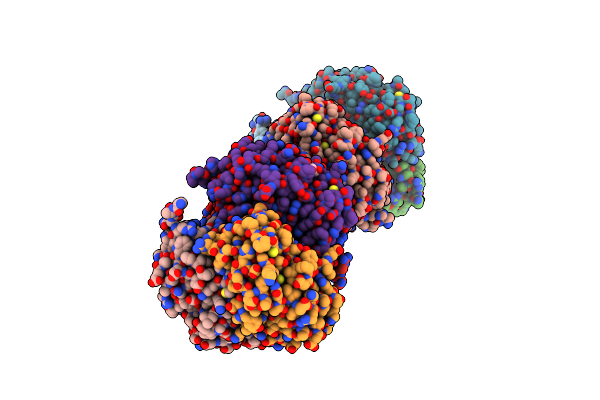
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: BTB, GOL |
 |
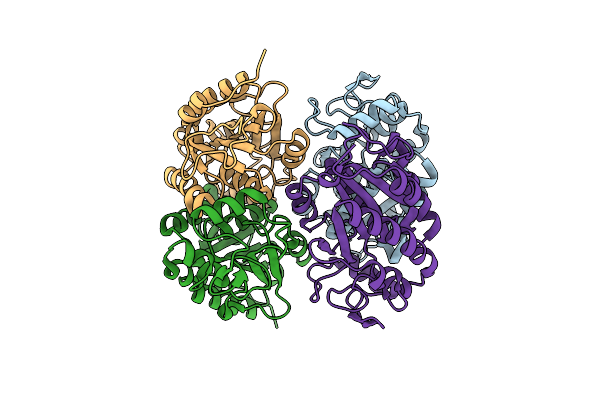
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: EDO, PO4 |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.46 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: CL |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:2.22 Å Release Date: 2018-02-21 Classification: HYDROLASE Ligands: C5J, CA |
 |
Structural And Biochemical Characterization Of A Non-Canonical Biuret Hydrolase (Biuh) From The Cyanuric Acid Catabolism Pathway Of Rhizobium Leguminasorum Bv. Viciae 3841
Organism: Rhizobium leguminosarum bv. viciae (strain 3841)
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2018-02-21 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: BTB, GOL, CL, C5S, PEG |
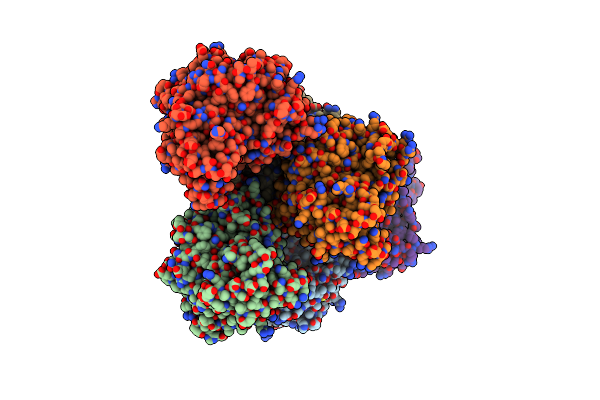 |
Structure Of The Amidase Domain Of Allophanate Hydrolase From Pseudomonas Sp Strain Adp
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2014-11-26 Classification: HYDROLASE Ligands: MLI |
 |
Structure Of Phosphotriesterase Mutant (S308L/Y309A) From Agrobacterium Radiobacter
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-09-24 Classification: HYDROLASE Ligands: FE2, CO, EDO |
 |
Structure Of Phosphotriesterase Mutant (S308L/Y309A) From Agrobacterium Radiobacter With Diethyl Thiophosphate Bound In The Active Site
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2014-09-24 Classification: HYDROLASE Ligands: FE2, CO, DPJ, EDO |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: PO4, MG, PEG |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, WDL, PEG |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation.
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, AX2 |
 |
Cyanuric Acid Hydrolase: Evolutionary Innovation By Structural Concatenation
Organism: Pseudomonas sp. adp
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2013-07-17 Classification: HYDROLASE Ligands: MG, BR8, PEG |
 |
Organism: Mycobacterium smegmatis str. mc2 155
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2010-04-14 Classification: FLAVOPROTEIN Ligands: EDO |

