Search Count: 42
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: FE2, FES, 8Q1 |
 |
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Frataxin-Bound)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: FE2, 8Q1 |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Fdx2-Bound In Proximal Conformation)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of The Core Isc Complex Under Turnover Conditions (Fdx2-Bound In Distal Conformation)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: TRANSFERASE Ligands: 8Q1, FE2, FES |
 |
Structure Of Human Cysteine Desulfurase Nfs1 With L-Propargylglycine Bound To Active Site Plp In Complex With Isd11, Acp1 And Iscu2
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-09-11 Classification: TRANSFERASE Ligands: EDO, PLP, LPH, GOL, PEG, PG4, P15, DTT, PGE, EDT, 8Q1, MES, 1PE |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Persulfide On Iscu2)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Persulfide On Nfs1 And Iscu2)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Structure Of The Human Mitochondrial Iron-Sulfur Cluster Biosynthesis Complex During Persulfide Transfer (Without Frataxin)
Organism: Homo sapiens, Escherichia coli bl21(de3)
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: TRANSFERASE Ligands: PLP, 8Q1, FE2 |
 |
Cryo-Em Structure Of The Electron Bifurcating Transhydrogenase Stnabc Complex From Sporomusa Ovata (State 2)
Organism: Sporomusa ovata dsm 2662
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: ELECTRON TRANSPORT Ligands: FES, ZN, FMN, SF4, NAD, FAD, NDP |
 |
Cryo-Em Structure Of The Electron Bifurcating Transhydrogenase Stnabc Complex From Sporomusa Ovata (State 1)
Organism: Sporomusa ovata dsm 2662
Method: ELECTRON MICROSCOPY Release Date: 2023-09-13 Classification: ELECTRON TRANSPORT Ligands: FES, ZN, SF4, FAD |
 |
Crystal Structure Of The Cerato-Platanin-Like Protein Cpl1 From Ustilago Maydis
Organism: Ustilago maydis 521
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-17 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Ustilago hordei
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2023-05-17 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, ZN, FMN, NAD |
 |
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Acetobacterium Woodii In The Reduced State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-22 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, HC1, ZN, FMN, NAI |
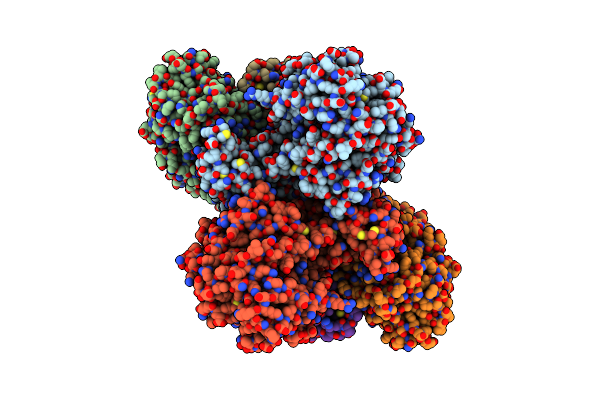 |
Cryoem Structure Of Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex A. Woodii In The Oxidised State
Organism: Acetobacterium woodii dsm 1030
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, ZN |
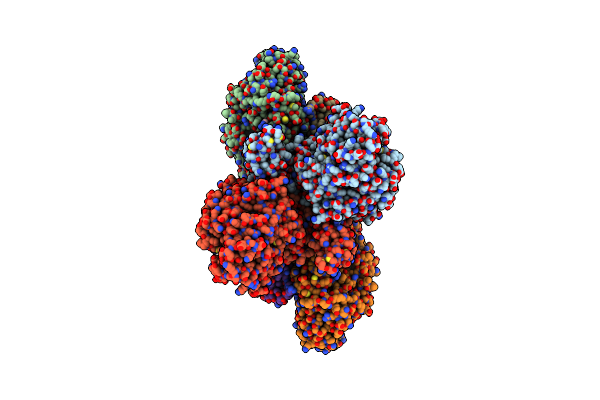 |
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Thermoanaerobacter Kivui In The Reduced State
Organism: Thermoanaerobacter kivui
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, HC1, FES, FMN, ZN, NAP |
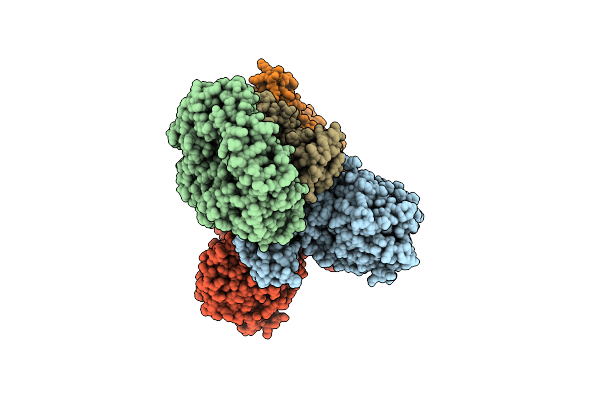 |
Cryo-Em Structure Of The Electron Bifurcating Fe-Fe Hydrogenase Hydabc Complex From Thermoanaerobacter Kivui In The Oxidised State
Organism: Thermoanaerobacter kivui
Method: ELECTRON MICROSCOPY Release Date: 2023-02-15 Classification: ELECTRON TRANSPORT Ligands: SF4, FES, FMN, ZN |
 |
Organism: Chaetomium thermophilum
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-09-21 Classification: PROTEIN BINDING Ligands: CIT |
 |
Cryo-Em Structure Of A Bacillus Subtilis Mifm-Stalled Ribosome-Nascent Chain Complex With (P)Ppgpp-Srp Bound
|

