Search Count: 82
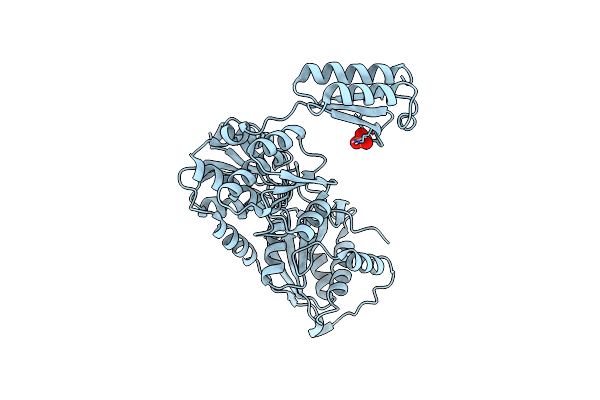 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.05 Å Release Date: 2021-10-27 Classification: ISOMERASE Ligands: PO4 |
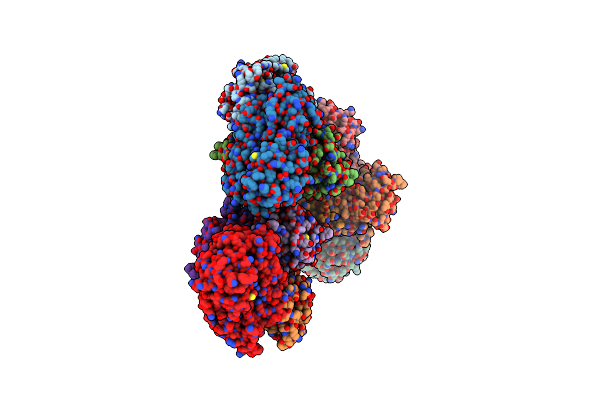 |
Complex Structure 2 Of The Bacillus Subtilis Cdaa C-Di-Amp Cyclase Domain (Cdaacd) And The Phosphoglucomutase Glmm Short Variant (Glmmf369)
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:4.20 Å Release Date: 2021-10-27 Classification: PROTEIN BINDING |
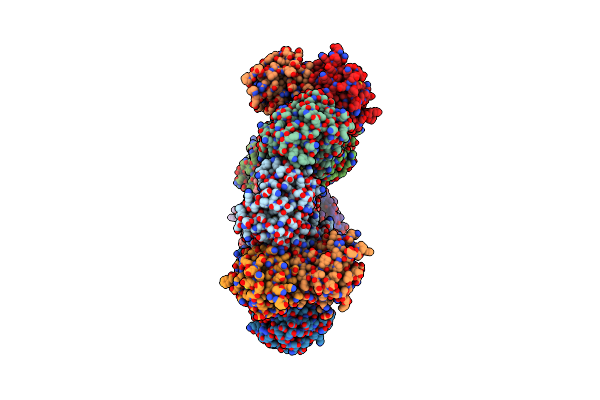 |
Bacillus Subtilis Complex Structure 1 Of Diadenylate Cyclase Cdaa Cytoplasmic Domain (Cdaacd) And The Phosphoglucomutase Glmm Short Variant (Glmmf369)
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:3.65 Å Release Date: 2021-10-27 Classification: PROTEIN BINDING |
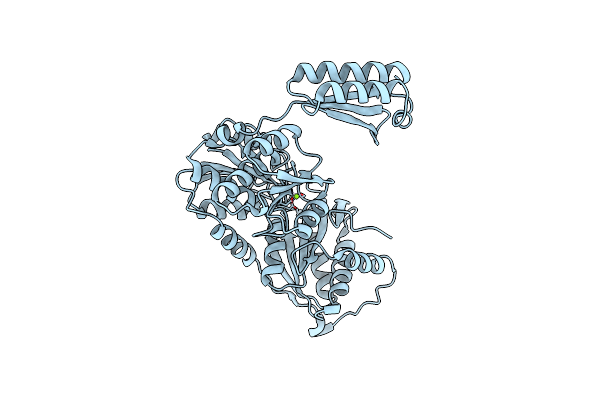 |
Organism: Bacillus subtilis (strain 168)
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2021-10-27 Classification: ISOMERASE Ligands: MG |
 |
Crystal Structure Of B. Subtilis Glucose-1-Phosphate Uridylyltransferase Yngb
Organism: Bacillus subtilis subsp. subtilis str. 168
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-02-10 Classification: TRANSFERASE |
 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-08 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia phage ms2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-08 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2019-07-24 Classification: METAL BINDING PROTEIN Ligands: ZN |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, ADP |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, ADP, NA, AWD |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, EJQ, ADP, NA |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, ADP, NA, ELQ |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, ADP, NA, EJW |
 |
Crystal Structure Of The P97 D2 Domain In A Helical Split-Washer Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.42 Å Release Date: 2019-04-10 Classification: HYDROLASE Ligands: MPD, DMS, ADP, NA, ELN |
 |
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 4-Fluoro-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, 4FW, MG |
 |
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 5-Methyl-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, D0Q, MG |
 |
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 6-Fluoro-L-Tryptophan
Organism: Chromobacterium violaceum atcc 12472
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, FT6, MG |
 |
Crystal Structure Of L-Tryptophan Oxidase Vioa From Chromobacterium Violaceum In Complex With 7-Methyl-L-Tryptophan
Organism: Chromobacterium violaceum (strain atcc 12472 / dsm 30191 / jcm 1249 / nbrc 12614 / ncimb 9131 / nctc 9757)
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2019-02-13 Classification: BIOSYNTHETIC PROTEIN Ligands: FAD, E95, MG |
 |
Structure Of 3' Phosphatase Nexo (Wt) From Neisseria Bound To Dna Substrate
Organism: Neisseria meningitidis serogroup b (strain mc58), Other sequences
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN Ligands: MPD |
 |
Structure Of 3' Phosphatase Nexo (D146N) From Neisseria Bound To Dna Substrate In Presence Of Magnesium Ion
Organism: Neisseria meningitidis mc58, Other sequences
Method: X-RAY DIFFRACTION Resolution:2.02 Å Release Date: 2018-10-31 Classification: DNA BINDING PROTEIN Ligands: MG, MPD |

