Search Count: 17
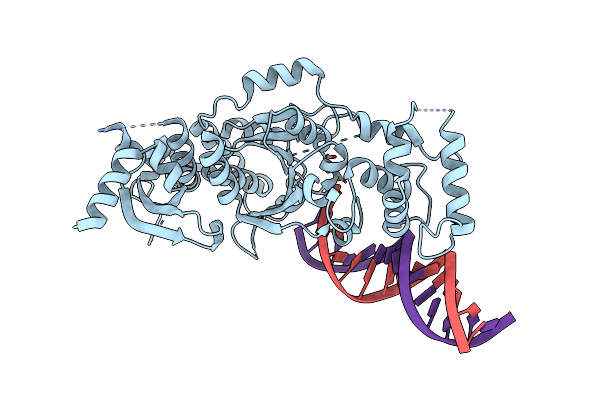 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-09-26 Classification: DNA Ligands: MG |
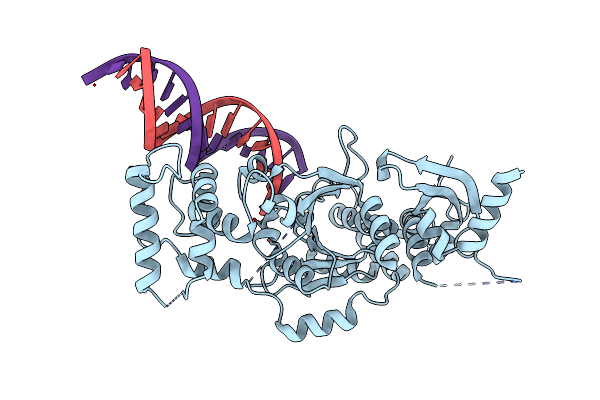 |
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Pichia kudriavzevii
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2018-09-26 Classification: DNA Ligands: MG |
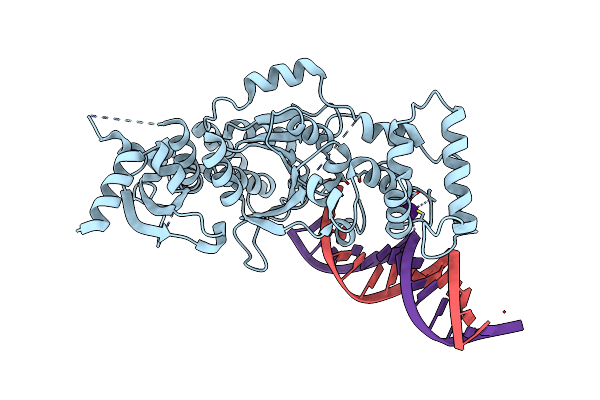 |
Organism: Chaetomium thermophilum, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2018-09-26 Classification: DNA Ligands: CS, MG |
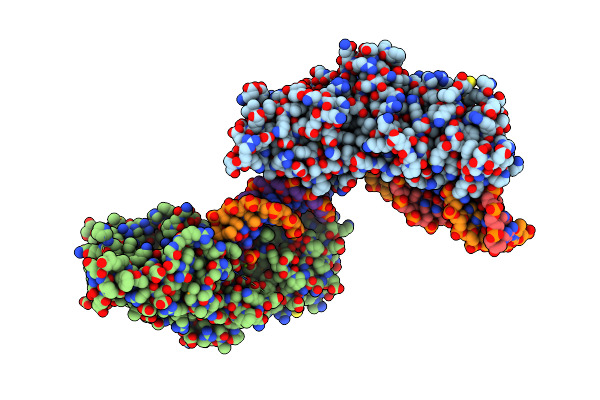 |
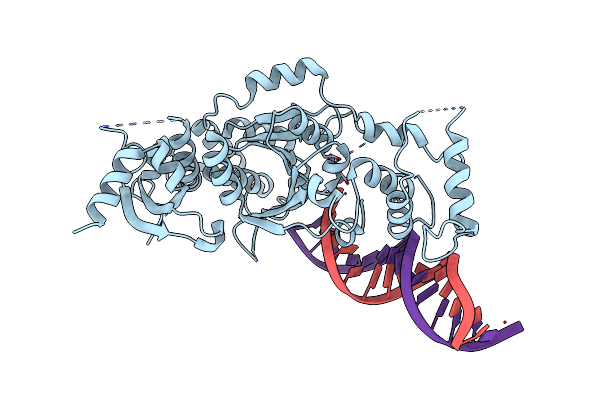
Crystal Structure Of The Holliday Junction-Resolving Enzyme Gen1 (Wt) In Complex With Product Dna And Mg2+ Ion
Organism: Chaetomium thermophilum (strain dsm 1495 / cbs 144.50 / imi 039719), Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-01-13 Classification: HYDROLASE Ligands: MG |
 |
Crystal Structure Of The Holliday Junction-Resolving Enzyme Gen1 (Wt) In Complex With Product Dna, Mg2+ And Mn2+ Ions
Organism: Chaetomium thermophilum, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-12-30 Classification: REPLICATION Ligands: MN |
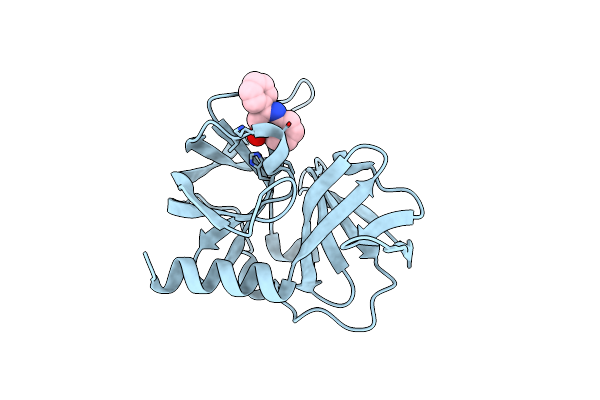 |
Organism: Human rhinovirus sp.
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-04-27 Classification: HYDROLASE Ligands: 7L4 |
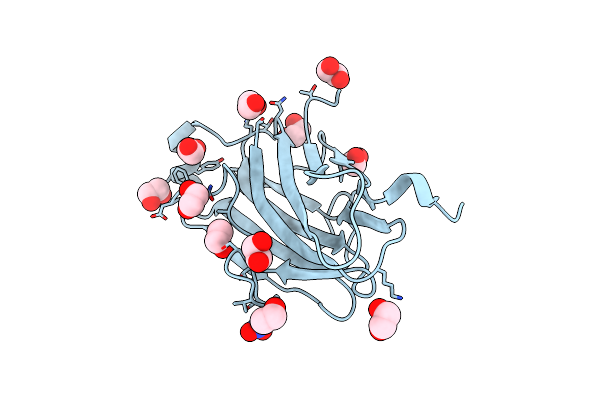 |
Crystal Structure Analysis Of The K171A Mutation Of N-Terminal Type Ii Cohesin 1 From The Cellulosomal Scab Subunit Of Acetivibrio Cellulolyticus
Organism: Acetivibrio cellulolyticus
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2008-12-02 Classification: STRUCTURAL PROTEIN Ligands: EDO, NH4, NO3 |
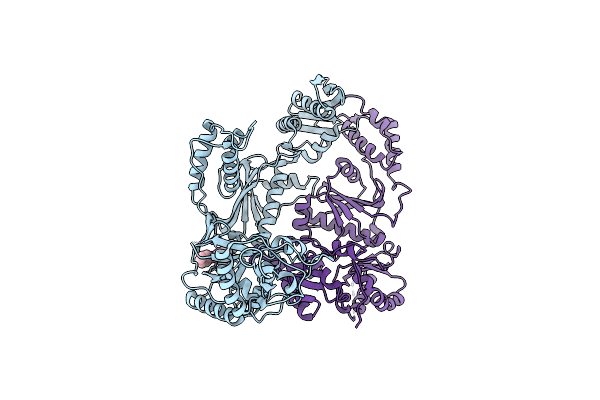 |
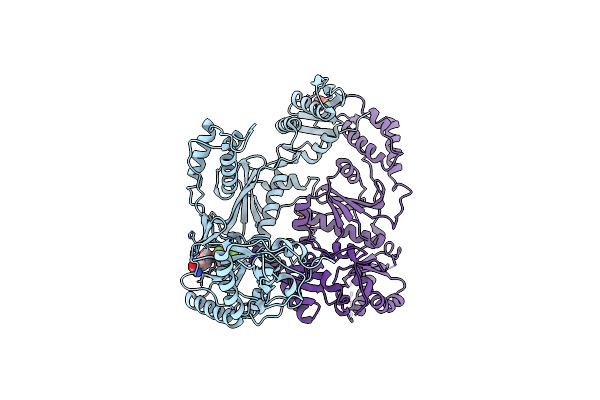
Organism: Hiv-1 m:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: GFA |
 |
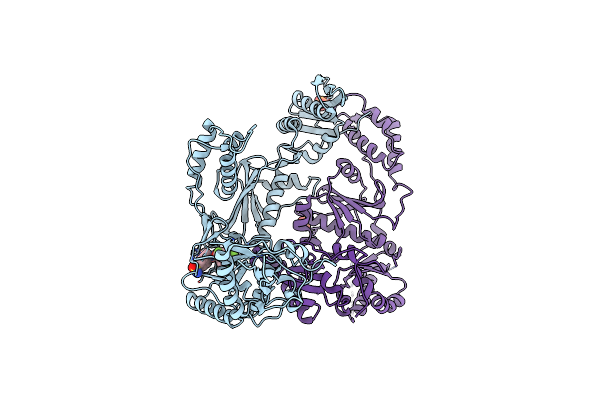
Organism: Hiv-1 m:b_hxb2r
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: PO4, GWE |
 |
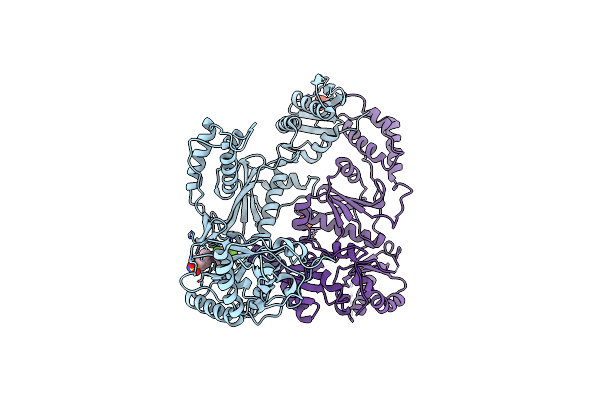
Crystal Structure Of Hiv-1 K103N Mutant Reverse Transcriptase In Complex With Gw564511.
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: PO4, GWE |
 |
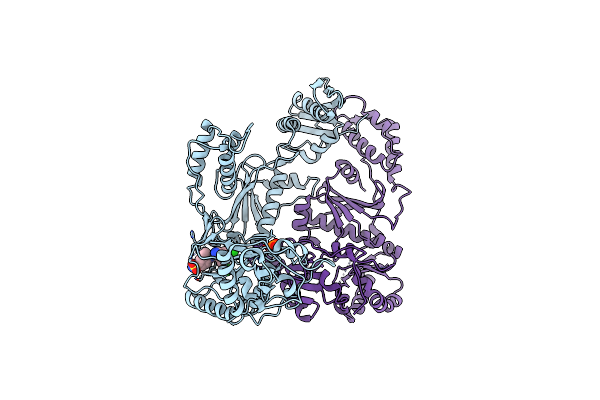
Crystal Structure Of Hiv-1 V106A And Y181C Mutant Reverse Transcriptase In Complex With Gw564511.
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: PO4, GWE |
 |
Crystal Structure Of K103N Mutant Hiv-1 Reverse Transcriptase In Complex With Gw678248.
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: PO4, GWJ |
 |
Crystal Structure Of L100I Mutant Hiv-1 Reverse Transcriptase In Complex With Gw695634.
Organism: Human immunodeficiency virus type 1
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-08-12 Classification: TRANSFERASE Ligands: PO4, GWI |
 |
Parent Structure Of Hen Egg White Lysozyme Grown In Acidic Ph 4.8. Refinement For Comparison With Crosslinked Molecules Of Lysozyme
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2007-07-24 Classification: HYDROLASE Ligands: CL, NA, NH4, EDO |
 |
Structure Of Hen Egg-White Lysozyme Determined From Crystals Grown In Ph 7.5
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2007-07-24 Classification: HYDROLASE |
 |
Crystal Structure Analysis Of Hen Egg White Lysozyme Crosslinked By Polymerized Glutaraldehyde In Acidic Environment
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2007-05-22 Classification: HYDROLASE Ligands: CL, NA, 220, EDO |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2007-05-22 Classification: HYDROLASE |

