Search Count: 56
 |
 |
 |
Focal Adhesion Kinase Catalytic Domain In Complex With 6-[4-(3-Methanesulfonyl-Benzylamino)-5-Trifluoromethyl-Pyrimidin-2-Ylamino]-3,4-Dihydro-1H-Quinolin-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: P4N |
 |
Focal Adhesion Kinase Catalytic Domain In Complex With N-Methyl-N-(3-{[2-(2-Oxo-1,2,3,4-Tetrahydro-Quinolin-6-Ylamino)-5-Trifluoromethyl-Pyrimidin-4-Ylamino]-Methyl}-Pyridin-2-Yl)-Methanesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: P7N, NA, SO4 |
 |
Focal Adhesion Kinase Catalytic Domain In Complex With N-Methyl-N-(2-{[2-(2-Oxo-2,3-Dihydro-1H-Indol-5-Ylamino)-5-Trifluoromethyl-Pyrimidin-4-Ylamino]-Methyl}-Phenyl)-Methanesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: P9K |
 |
Focal Adhesion Kinase Catalytic Domain In Complex With N-Methyl-N-(3-{[2-(2-Oxo-2,3-Dihydro-1H-Indol-5-Ylamino)-Pyrimidin-4-Ylamino]-Methyl}-Pyridin-2-Yl)-Methanesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: PKE, SO4 |
 |
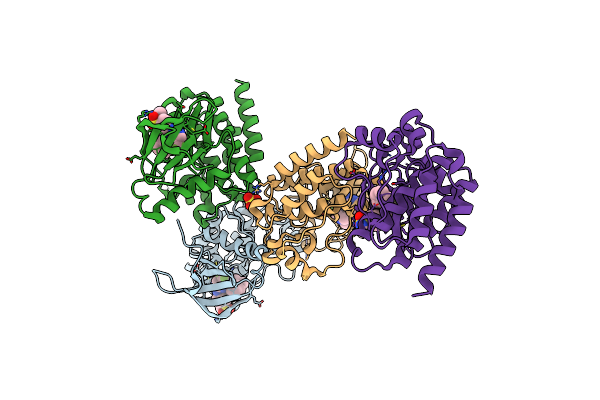
Focal Adhesion Kinase Catalytic Domain In Complex With 5-{4-[(Pyridin-3-Ylmethyl)-Amino]-5-Trifluoromethyl-Pyrimidin-2-Ylamino}-1,3-Dihydro-Indol-2-One
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: PVT, SO4 |
 |
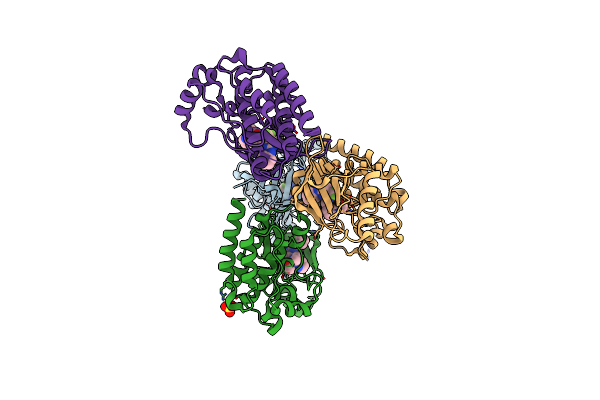
Focal Adhesion Kinase Catalytic Domain In Complex With 4-{[4-{[(1R,2R)-2-(Dimethylamino)Cyclopentyl]Amino}-5-(Trifluoromethyl)Pyrimidin-2-Yl]Amino}-N-Methylbenzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: P1E, SO4 |
 |
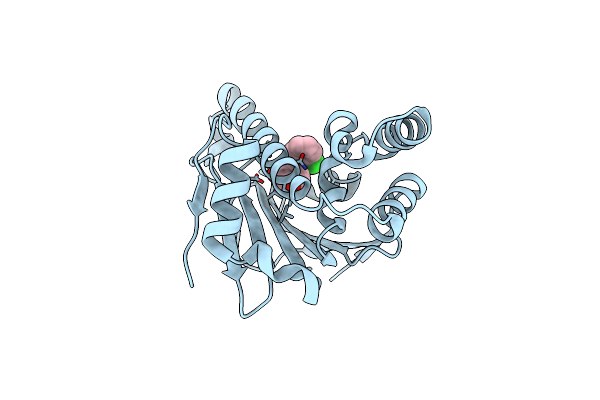
Focal Adhesion Kinase Catalytic Domain In Complex With N-Methyl-N-{3-[(2-Phenylamino-5-Trifluoromethyl-Pyrimidin-4-Ylamino)-Methyl]-Pyridin-2-Yl}-Methanesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-02-10 Classification: TRANSFERASE Ligands: Q2H, SO4 |
 |
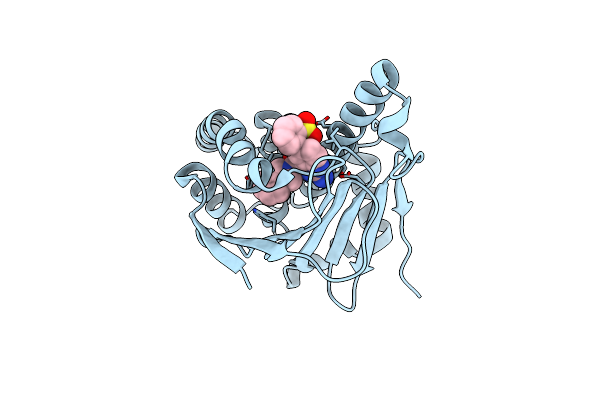
Estimation Of Protein-Ligand Unbinding Kinetics Using Non-Equilibrium Targeted Molecular Dynamics Simulations
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2019-01-30 Classification: CHAPERONE Ligands: D4W |
 |
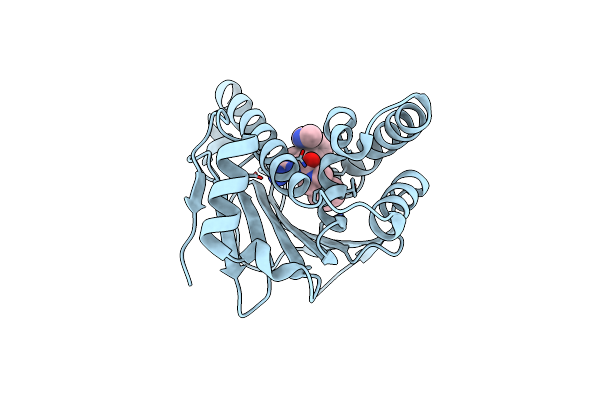
Hsp90 Inhibitor Desolvation As A Rationale To Steer On-Rates And Impact Residence Time
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-11-21 Classification: CHAPERONE Ligands: H0T |
 |
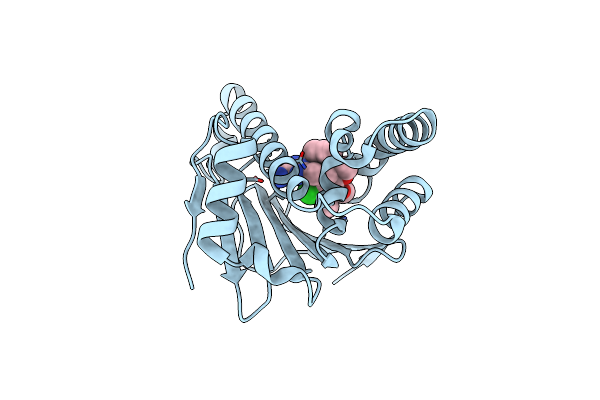
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: B5Q |
 |
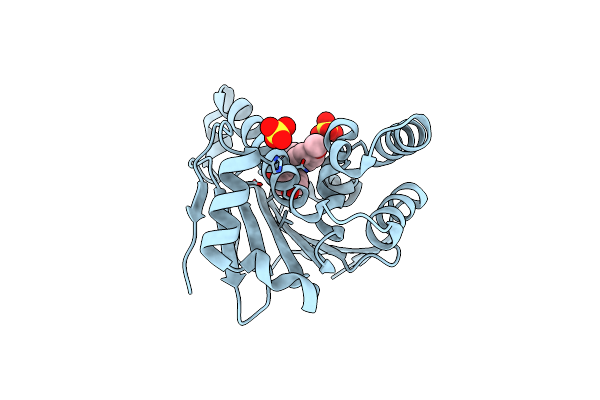
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: PU1 |
 |
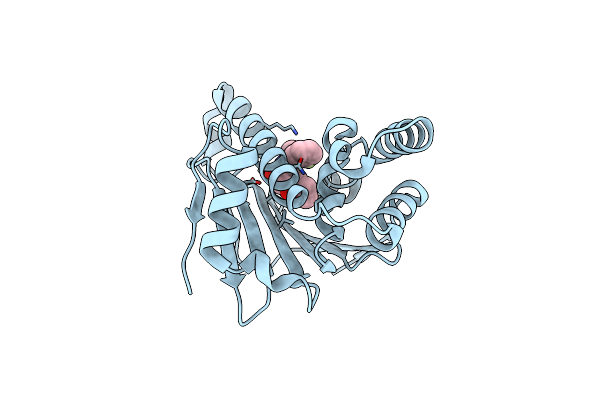
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: P4A, SO4 |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: BAW |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: BA8 |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: DMS, C4T, SO4 |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: SO4, C4N |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: C4K |
 |
Estimation Of Relative Drug-Target Residence Times By Random Acceleration Molecular Dynamics Simulation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: CHAPERONE Ligands: C3Z, SO4 |

