Search Count: 65
 |
Crystal Structure Of The Dyp-Type Peroxidase Pross Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: OXY, HEM, CL |
 |
Crystal Structure Of A Dyp-Type Peroxidase Fireprot Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-04-30 Classification: OXIDOREDUCTASE Ligands: GOL, HEM, CL |
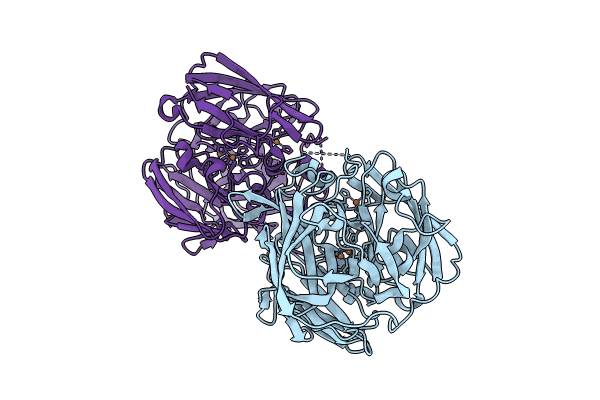 |
Crystal Structure Of A Multicopper Oxidase 3F3 Variant From Pyrobaculum Aerophilum
Organism: Pyrobaculum aerophilum str. im2
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2024-02-21 Classification: OXIDOREDUCTASE Ligands: CU, CL |
 |
Crystal Structure Determination Of Dye-Decolorizing Peroxidase (Dyp) From Deinoccoccus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN Ligands: HEM, GOL, MG |
 |
Crystal Structure Determination Of Dye-Decolorizing Peroxidase (Dyp) Mutant M190G From Deinoccoccus Radiodurans
Organism: Deinococcus radiodurans
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-01-31 Classification: STRUCTURAL PROTEIN Ligands: HEM, OH, CL, CA |
 |
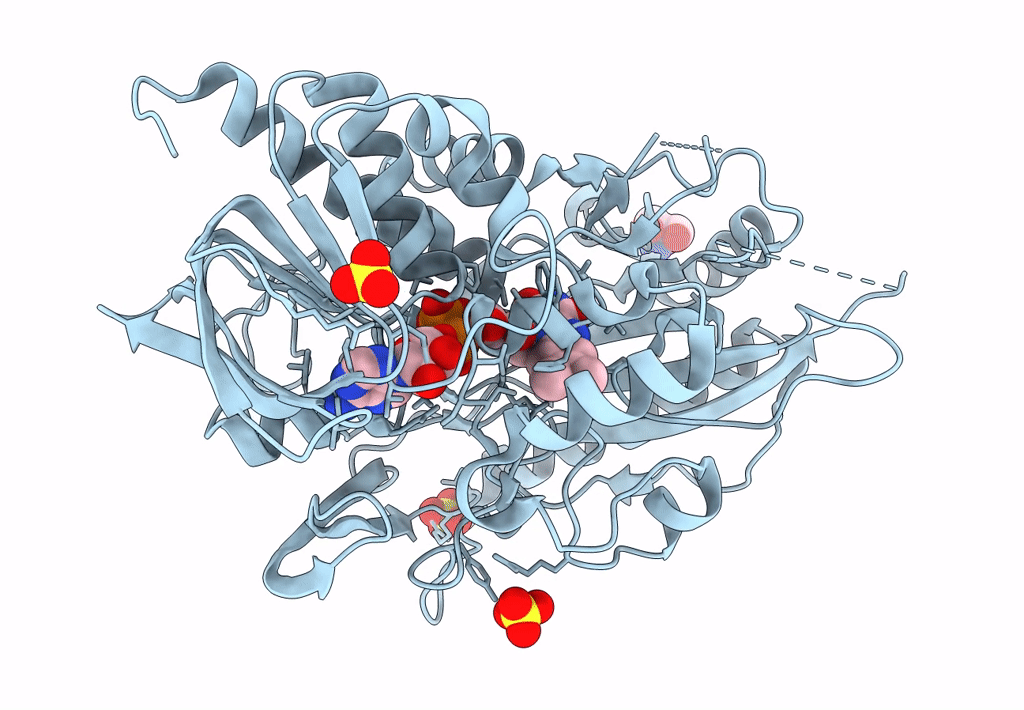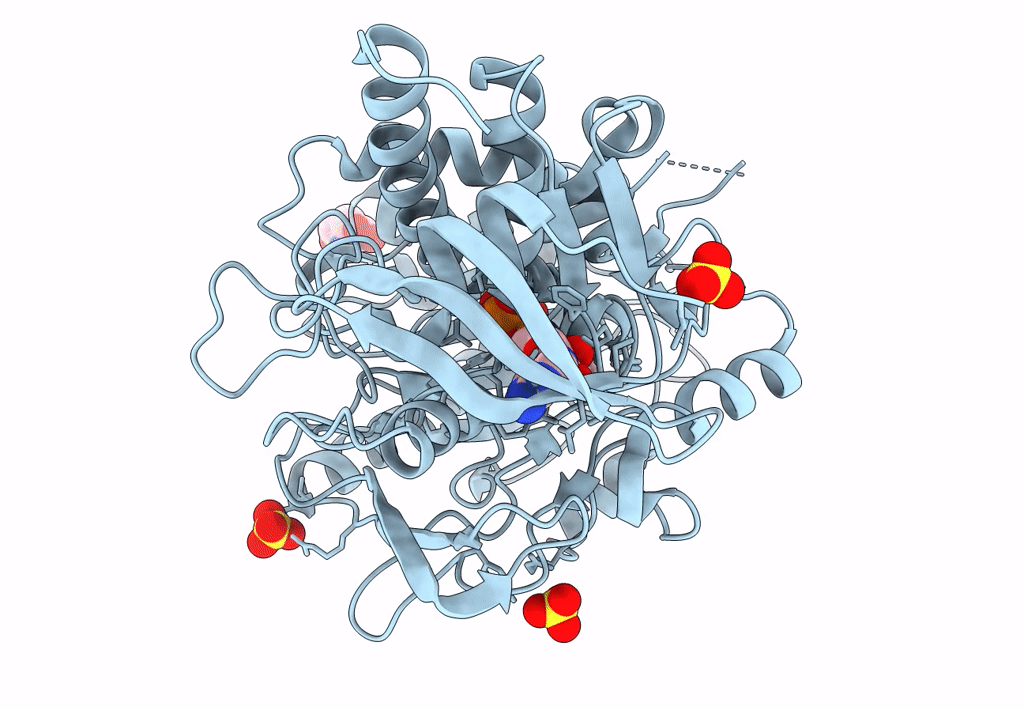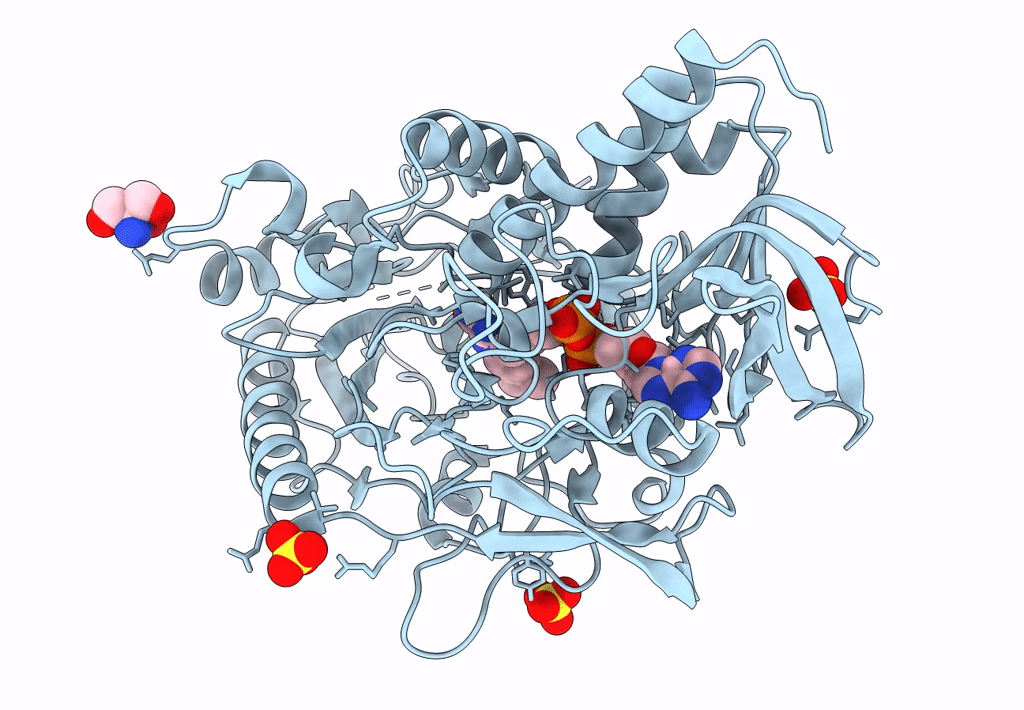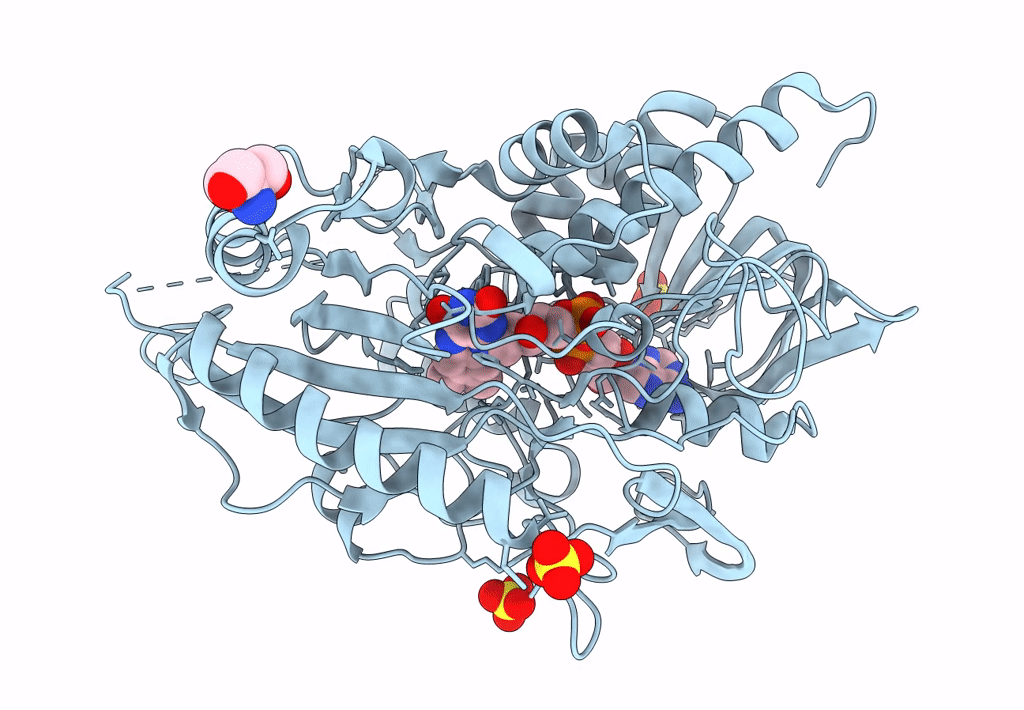
Crystal Structure Of A Bacterial Pyranose 2-Oxidase In Complex With Mangiferin
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2023-08-16 Classification: OXIDOREDUCTASE Ligands: FAD, HZI, SO4 |
 |
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: FAD, GLC, SO4, TRS |
 |
Crystal Structure Of A Bacterial Pyranose 2-Oxidase From Pseudoarthrobacter Siccitolerans
Organism: Pseudarthrobacter siccitolerans
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2023-06-14 Classification: OXIDOREDUCTASE Ligands: SO4, TRS, FAD |
 |
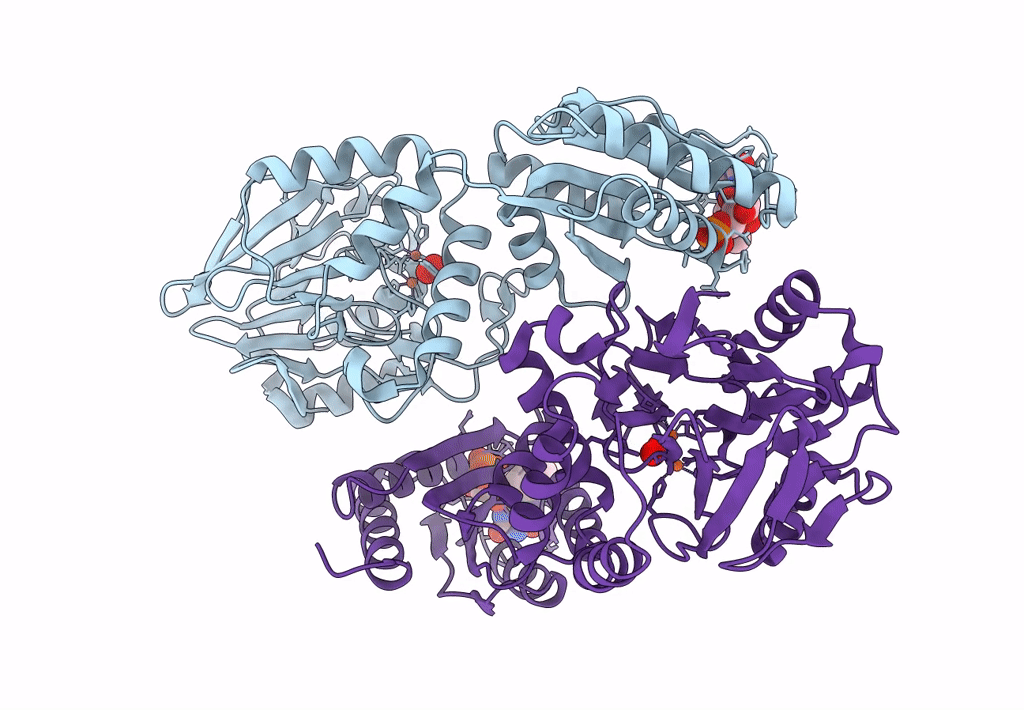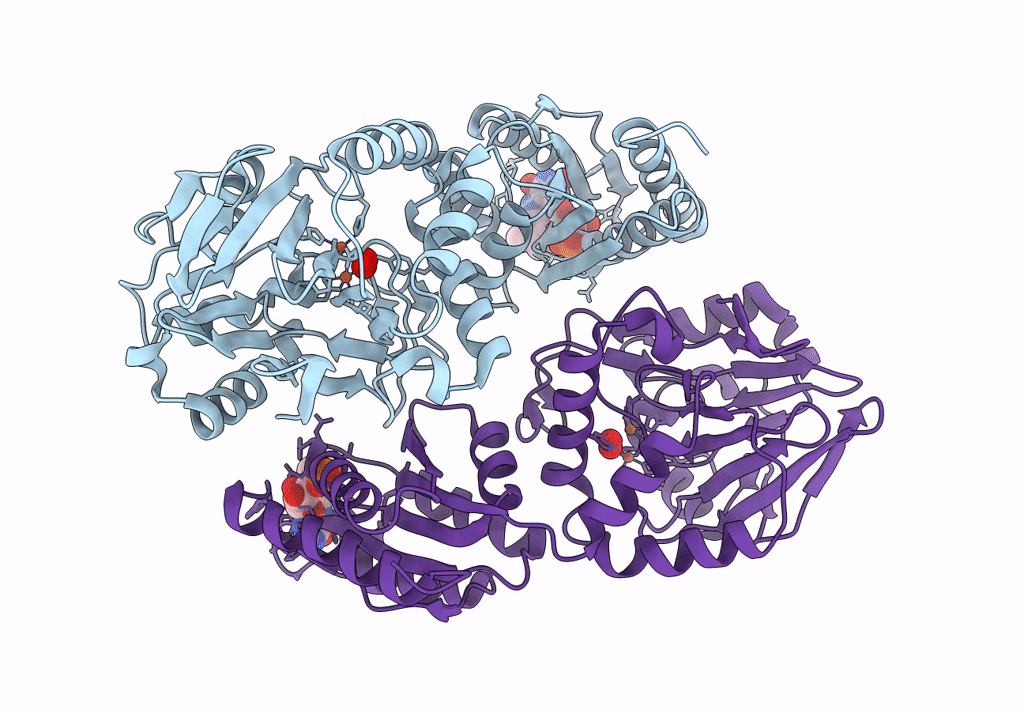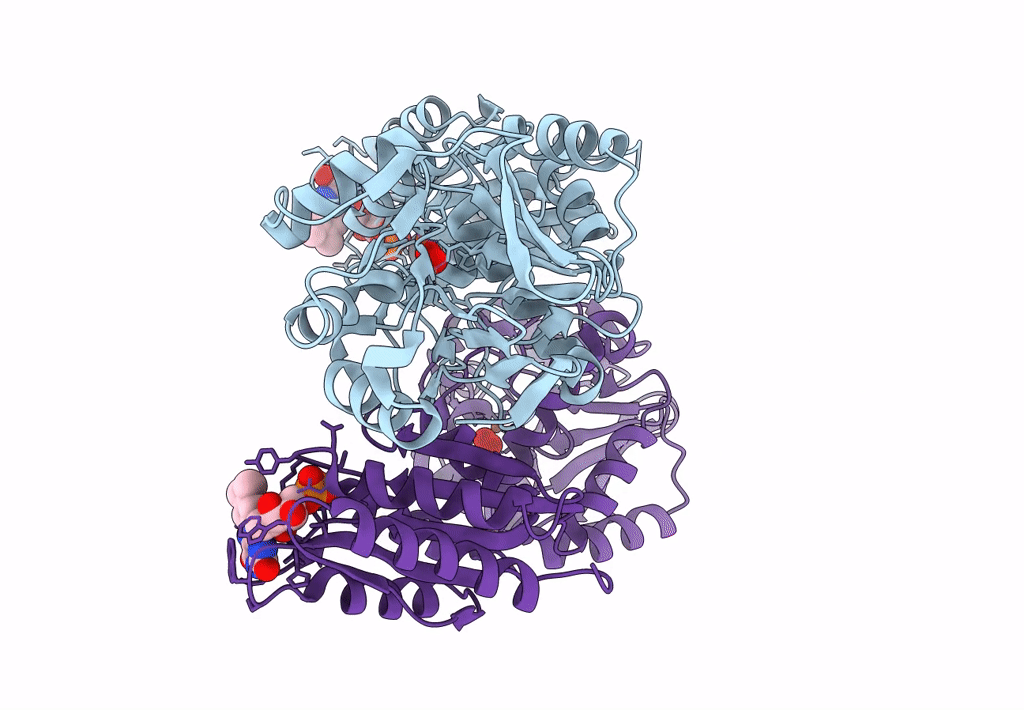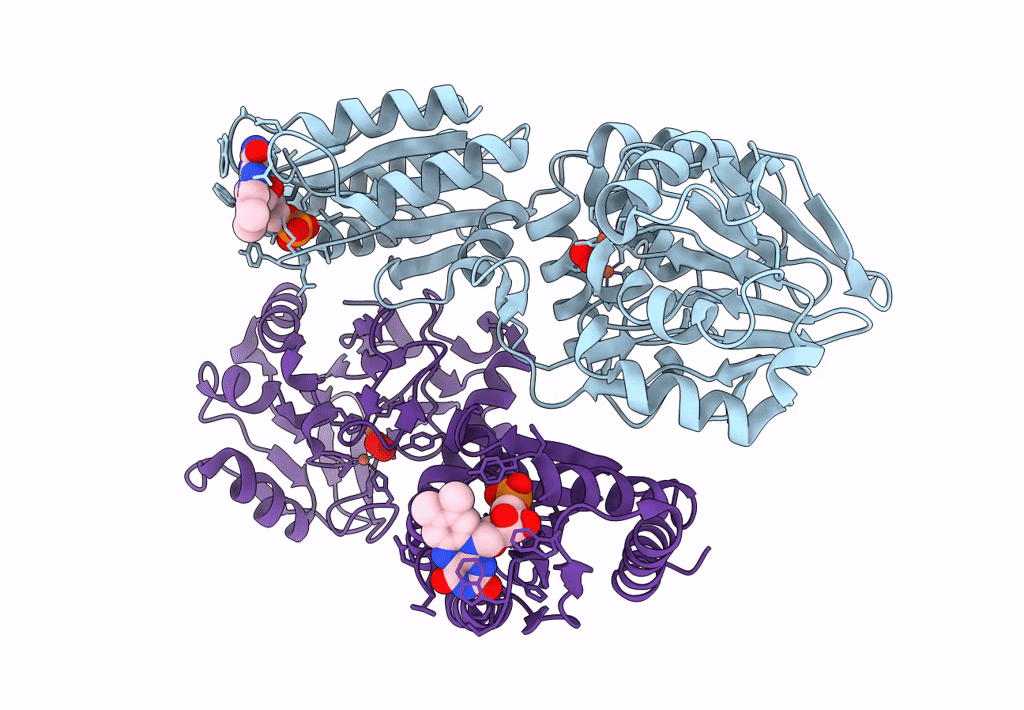
Crystal Structure Of A Flavodiiron Protein D52K Mutant In The Oxidized State From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, OXY |
 |
Crystal Structure Of A Flavodiiron Protein D52K Mutant In The Reduced State From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, OXY, IPA |
 |
Crystal Structure Of A Flavodiiron Protein S262Y Mutant In The Reduced State From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, OXY |
 |
Crystal Structure Of A Flavodiiron Protein S262Y Mutant In The Oxidized State From Escherichia Coli
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FEO, GOL |
 |
Crystal Structure Of A Flavodiiron Protein D52K/S262Y Mutant In The Oxidized State From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, IPA, OXY, O |
 |
Crystal Structure Of A Flavodiiron Protein D52K/S262Y Mutant In The Reduced State From Escherichia Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, OXY |
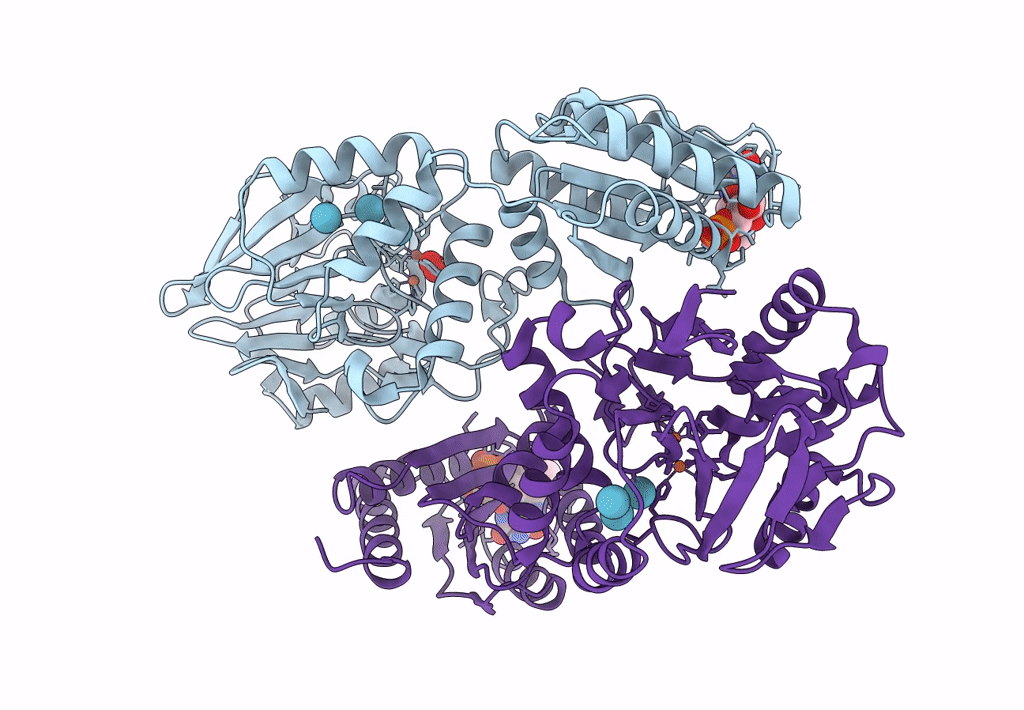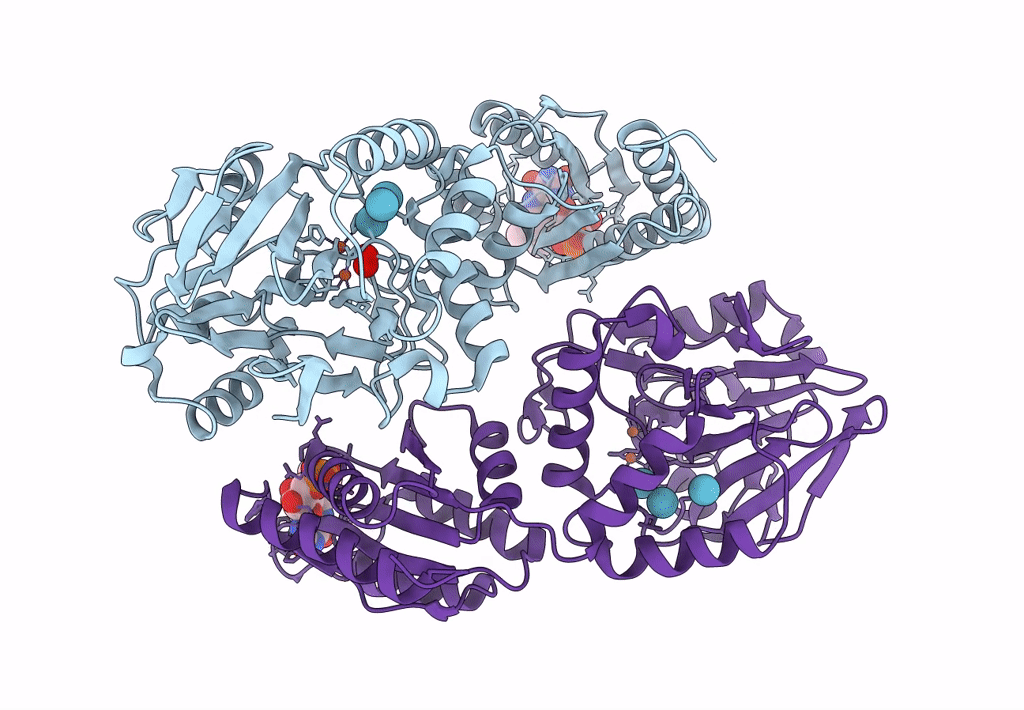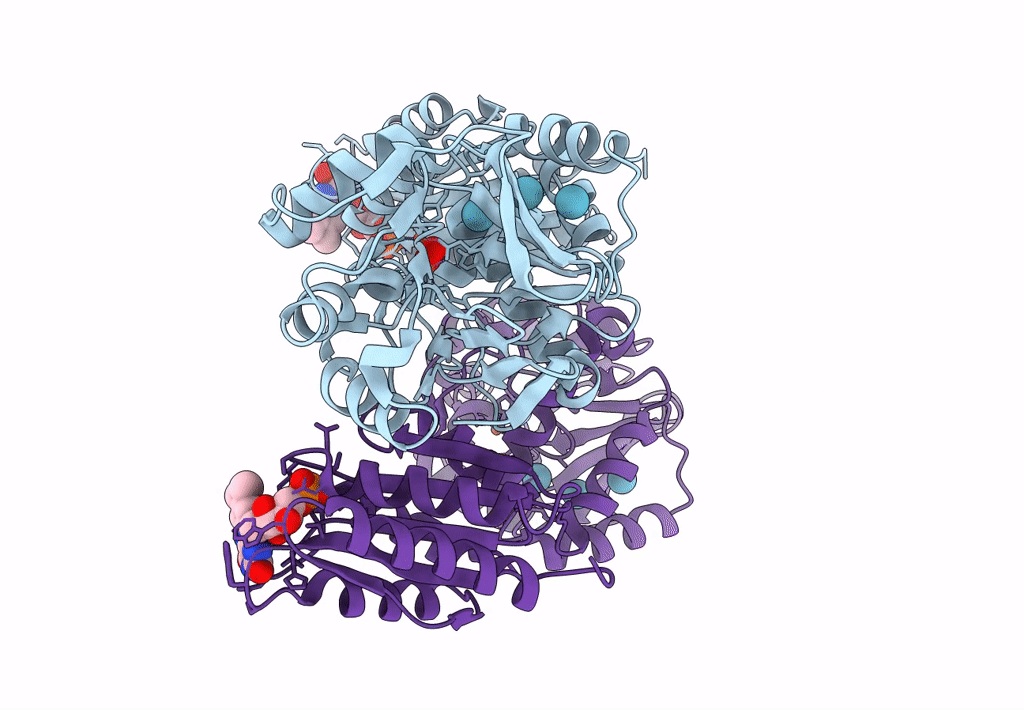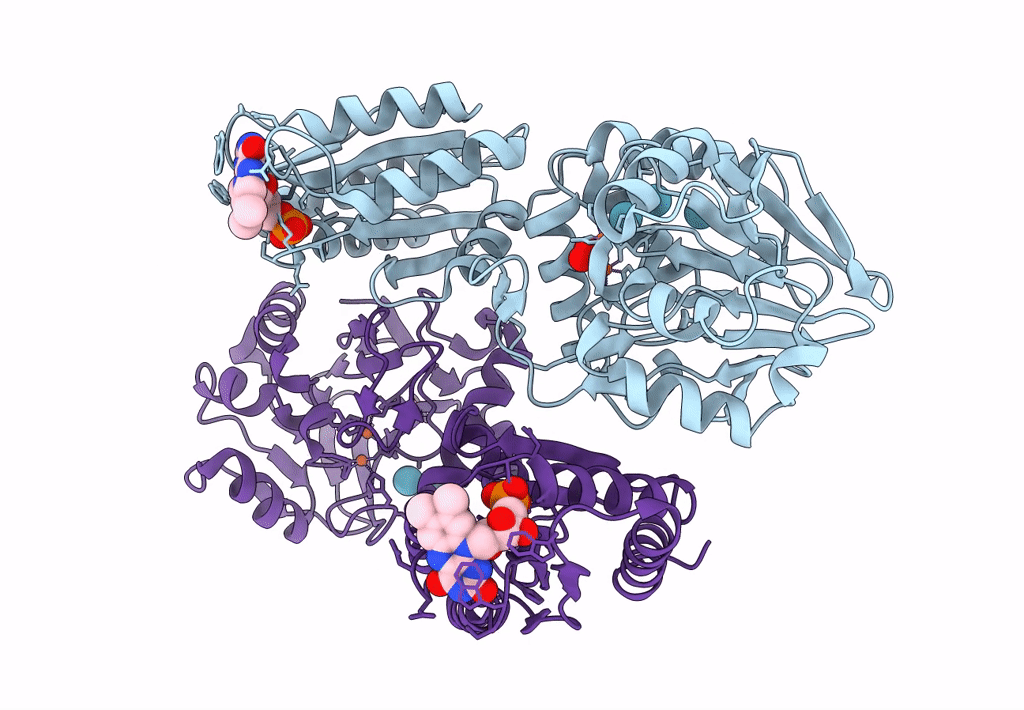 |
Crystal Structure Of A Flavodiiron Protein D52K Mutant From Escherichia Coli Pressurized With Krypton Gas
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2023-02-15 Classification: OXIDOREDUCTASE Ligands: FMN, FE, O, KR, OXY |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-08-03 Classification: OXIDOREDUCTASE Ligands: HEM |
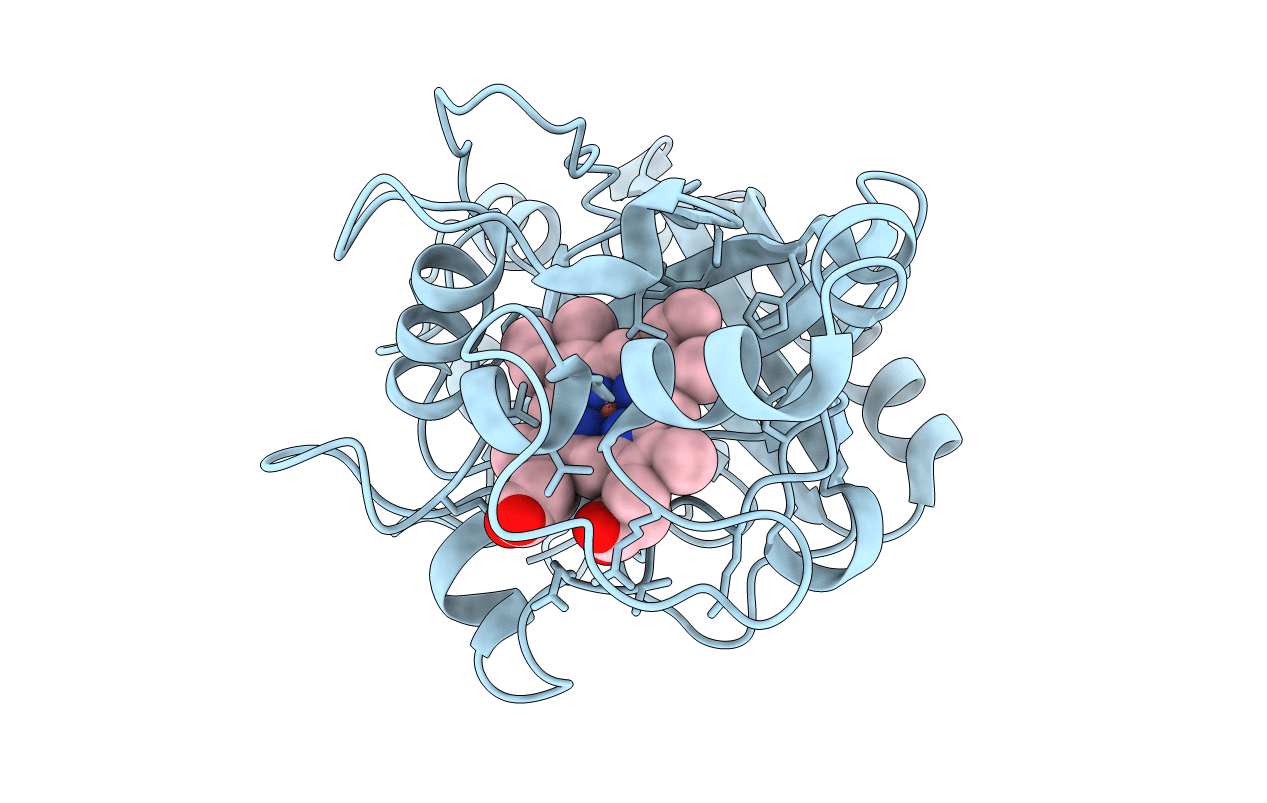 |
Crystal Structure Of A Dyp-Type Peroxidase 6E10 Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2022-08-03 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of A Dyp-Type Peroxidase 29E4 Variant From Pseudomonas Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-08-03 Classification: OXIDOREDUCTASE Ligands: HEM |
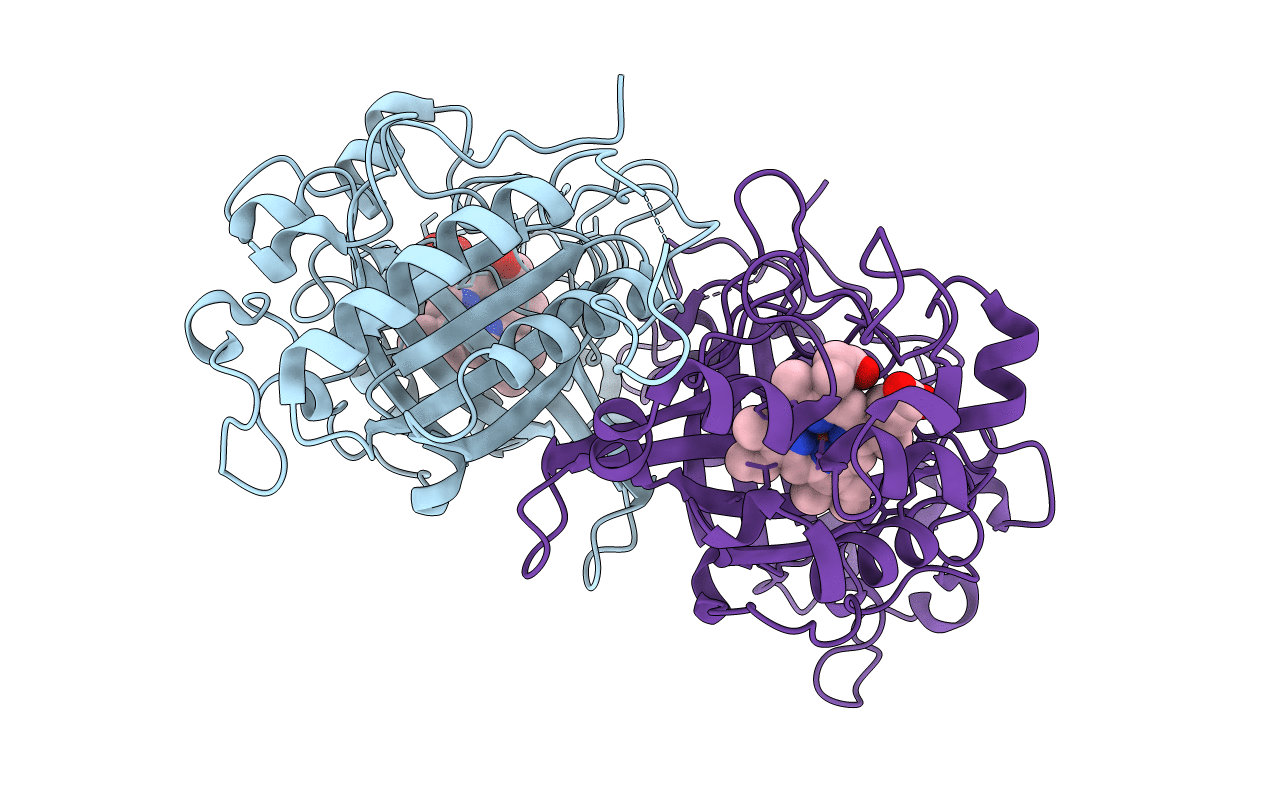 |
Crystal Structure Of A Dyp-Type Peroxidase From Bacillus Subtilis In P3121 Space Group
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2021-10-27 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
Crystal Structure Of A Dyp-Type Peroxidase 5G5 Variant From Bacillus Subtilis
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-10-27 Classification: OXIDOREDUCTASE Ligands: HEM |

