Search Count: 18
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
Crystal Structure Of Ctag From Ruminiclostridium Cellulolyticum (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: GOL |
 |
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-10-01 Classification: LIGASE |
 |
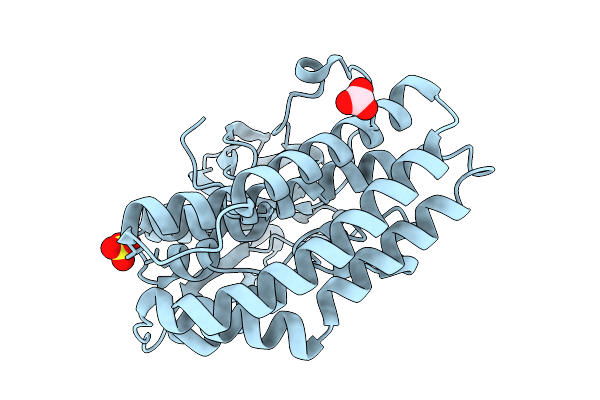
Crystal Structure Of The Ctag_C11A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: NI, IMD, TRS |
 |
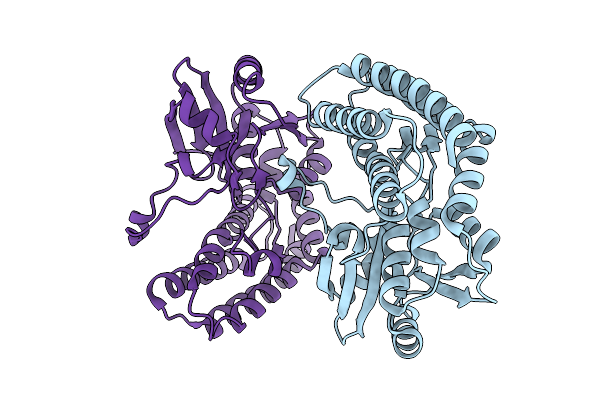
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: SO4, CO3 |
 |
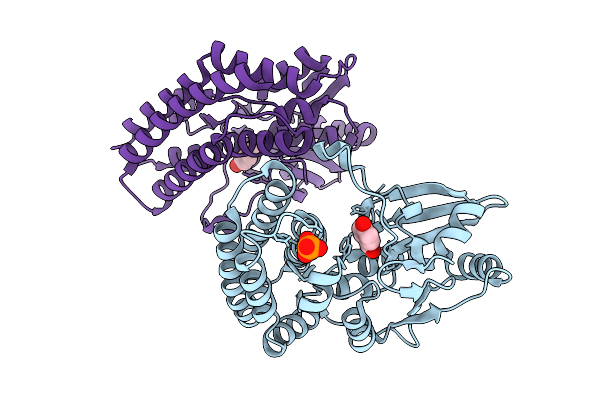
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-10-01 Classification: LIGASE |
 |
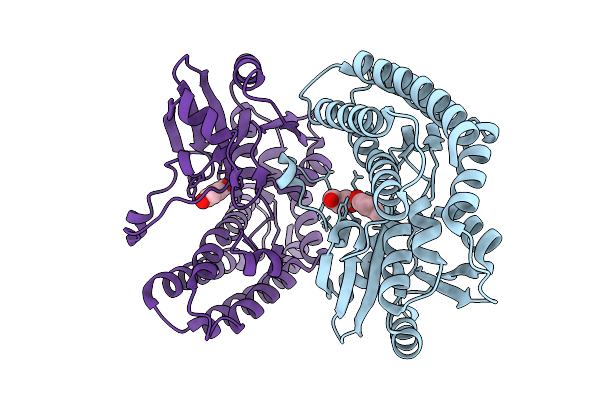
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, PO4 |
 |
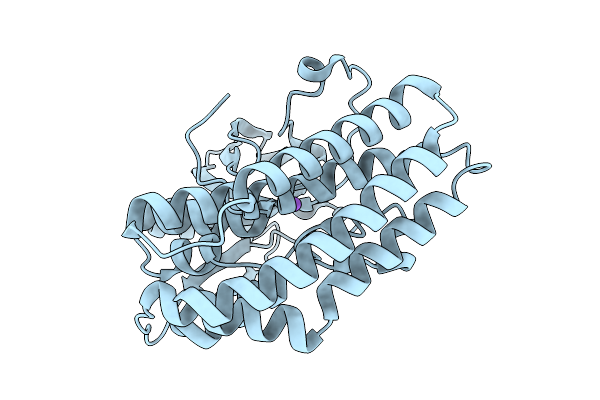
Crystal Structure Of The Ctag_H128A Variant From Ruminiclostridium Cellulolyticum In Complex With Phba (P2(1)2(1)2(1)-Medium)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-10-01 Classification: LIGASE Ligands: HBA, GOL |
 |
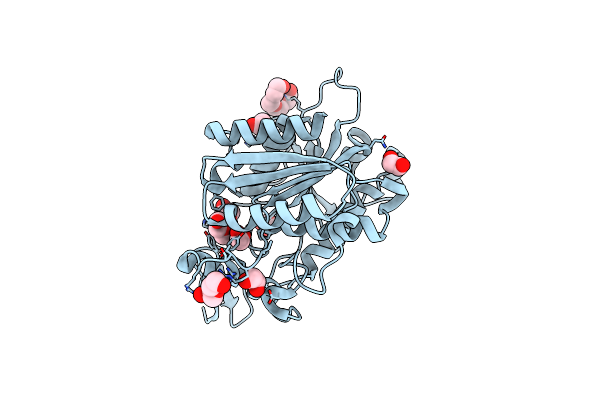
Crystal Structure Of The Ctag_D144N Variant From Ruminiclostridium Cellulolyticum (P2(1)-Small)
Organism: Ruminiclostridium cellulolyticum
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2025-10-01 Classification: LIGASE |
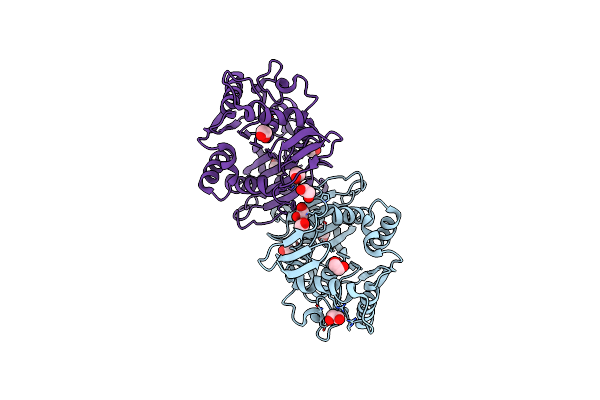
 |
Organism: Tabernanthe iboga
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: PG4, PEG, EDO |
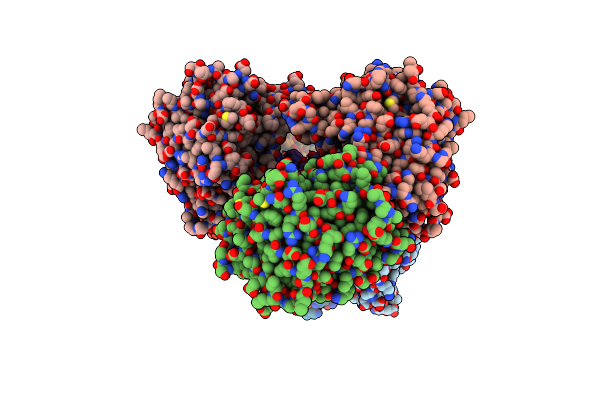
 |
Structure Of Tabersonine Synthase - An Alpha-Beta Hydrolase From Catharanthus Roseus
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: EDO |
 |
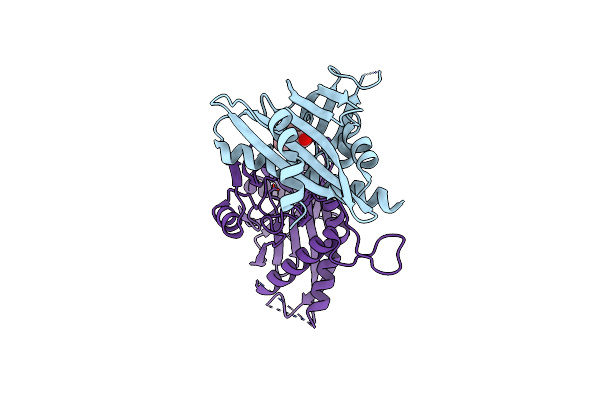
Structure Of Catharanthine Synthase - An Alpha-Beta Hydrolase From Catharanthus Roseus With A Cleaviminium Intermediate Bound
Organism: Catharanthus roseus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2020-03-04 Classification: HYDROLASE Ligands: KJE, P6G |
 |
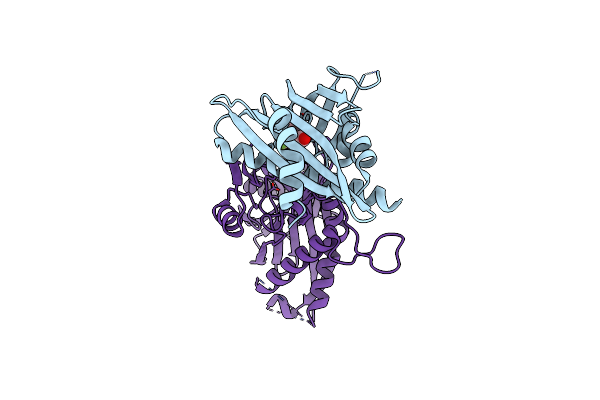
Crystal Structures Of Pyr1/Hab1 In Complex With Synthetic Analogues Of Abscisic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-06-27 Classification: PROTEIN BINDING Ligands: A4H, MN |
 |
Crystal Structures Of Pyr1/Hab1 In Complex With Synthetic Analogues Of Abscisic Acid
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2018-06-27 Classification: PROTEIN BINDING Ligands: A4K, MN |
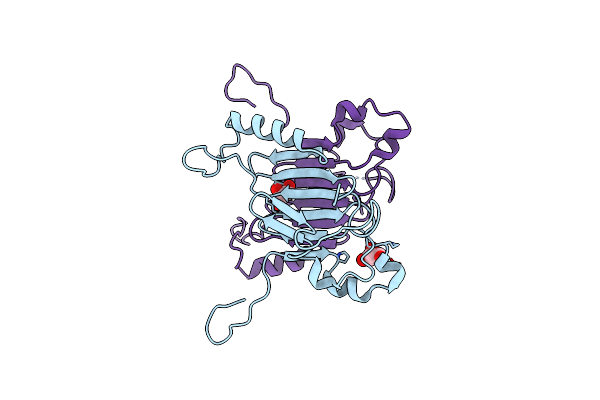 |
Crystal Structure Of The C-Terminal Domain Of Nuclear Pore Complex Component Nup116 From Candida Glabrata
Organism: Candida glabrata
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-08-04 Classification: PROTEIN TRANSPORT Ligands: GOL |
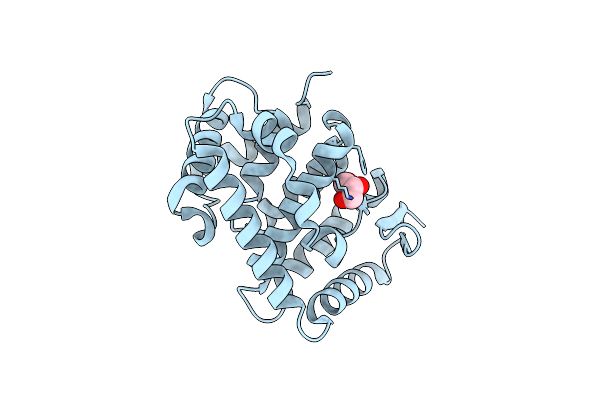 |
Crystal Structure Of The C-Terminal Domain From The Nuclear Pore Complex Component Nup133 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-01-26 Classification: PROTEIN TRANSPORT Ligands: GOL |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of The Autoproteolytic Domain From The Nuclear Pore Complex Component Nup145 From Saccharomyces Cerevisiae In The Hexagonal, P61 Space Group
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2009-12-22 Classification: PROTEIN TRANSPORT, RNA BINDING PROTEIN Ligands: EDO |

