Search Count: 32
 |
Organism: Human coronavirus oc43
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: A1AUY |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: A1AUX |
 |
Co-Structure Of Main Protease Of Sars-Cov-2 (Covid-19) With Covalent Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-10-16 Classification: HYDROLASE Ligands: A1AU4 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: PO4, DMS, O5J |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 2
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5P |
 |
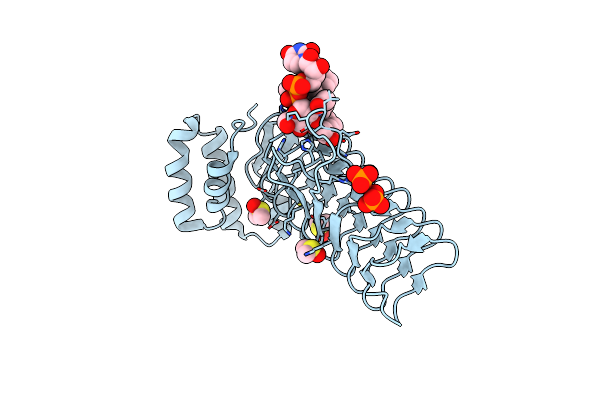
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 6
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5M |
 |
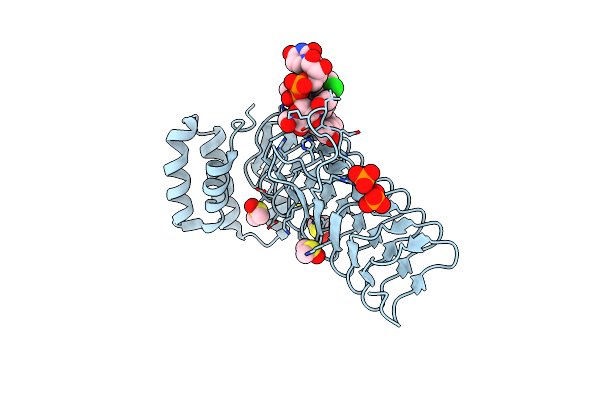
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 7
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5G |
 |
E.Coli Lpxa In Complex With Udp-3-O-(R-3-Hydroxymyristoyl)-Glcnac And Compound 8
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2020-03-11 Classification: TRANSFERASE/INHIBITOR Ligands: U20, PO4, DMS, O5D |
 |
Structure Of The Human Ddb1-Dda1-Dcaf15 E3 Ubiquitin Ligase Bound To Rbm39 And Indisulam
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-18 Classification: ONCOPROTEIN Ligands: EF6 |
 |
Crystal Structure Of Full-Length Human Dcaf15-Ddb1(Deltabpb)-Dda1-Rbm39 In Complex With Indisulam
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-12-18 Classification: LIGASE Ligands: GOL, EF6 |
 |
Crystal Structure Of Full-Length Human Dcaf15-Ddb1-Deltapbp-Dda1-Rbm39 In Complex With 4-(Aminomethyl)-N-(3-Cyano-4-Methyl-1H-Indol-7-Yl)Benzenesulfonamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.61 Å Release Date: 2019-12-18 Classification: LIGASE Ligands: Q5J, GOL |
 |
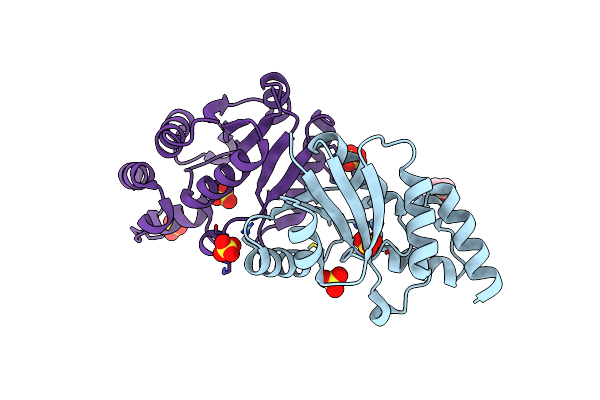
Crystal Structure Of A Zinc-Binding Non-Structural Protein From The Hepatitis E Virus
Organism: Hepatitis e virus
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2019-02-13 Classification: VIRAL PROTEIN Ligands: ZN, NO2, GOL |
 |
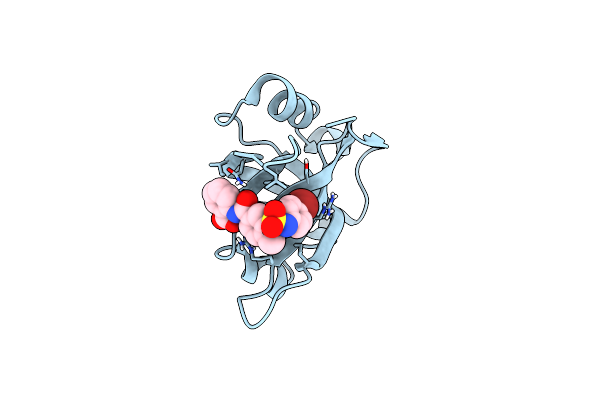
Crystal Structure Of Apo Phosphopantetheine Adenylyltransferase (Ppat/Coad) From E. Coli
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2016-05-25 Classification: TRANSFERASE Ligands: SO4, DMS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.21 Å Release Date: 2016-01-27 Classification: PROTEIN BINDING/INHIBITOR Ligands: 5KR |
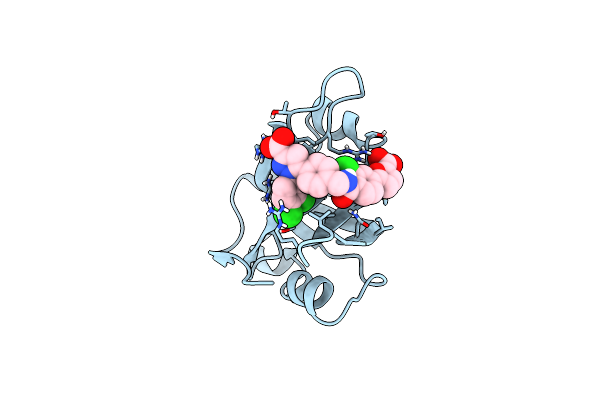 |
Structure Of Rpa70N In Complex With 5-(4-((4-(5-Carboxyfuran-2-Yl)-2-Chlorobenzamido)Methyl)Phenyl)-1-(3,4-Dichlorophenyl)-1H-Pyrazole-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: 3HS |
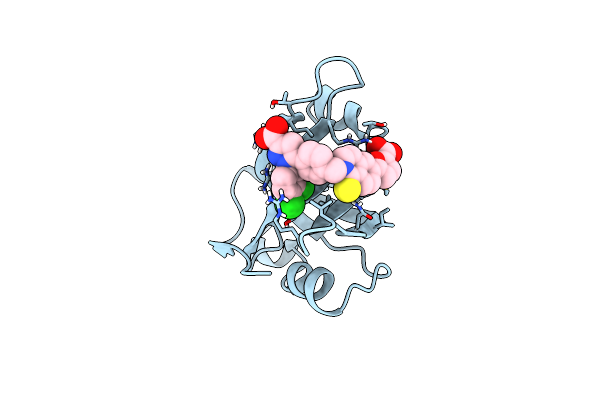 |
Structure Of Rpa70N In Complex With 5-(4-((6-(5-Carboxyfuran-2-Yl)-1-Thioxo-3,4-Dihydroisoquinolin-2(1H)-Yl)Methyl)Phenyl)-1-(3,4-Dichlorophenyl)-1H-Pyrazole-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: 3HV |
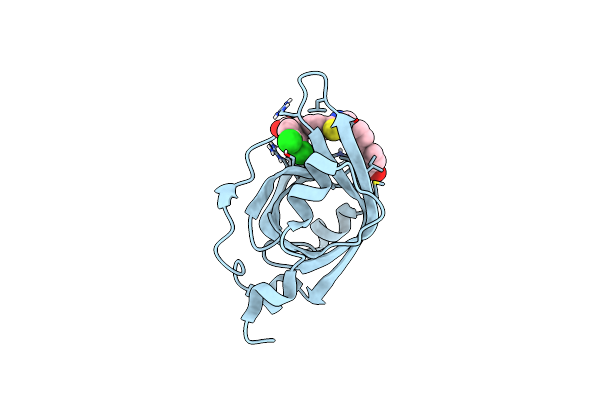 |
Crystal Structure Of Rpa70N In Complex With 5-(4-((4-(5-Carboxyfuran-2-Yl)Benzyl)Carbamothioyl)Phenyl)-1-(3,4-Dichlorophenyl)-1H-Pyrazole-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: 3HW |
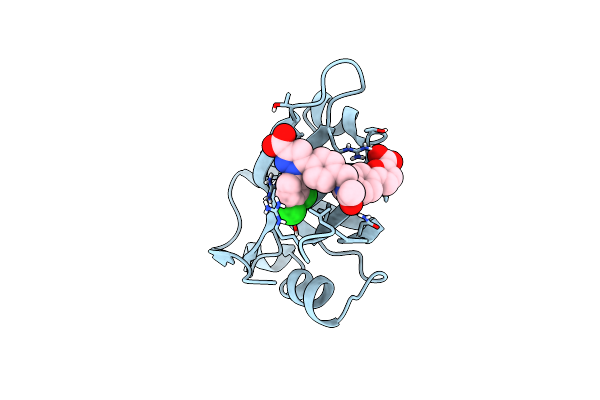 |
Crystal Structure Of Rpa70N In Complex With 5-(3-((N-(4-(5-Carboxyfuran-2-Yl)Benzyl)Acetamido)Methyl)Phenyl)-1-(3,4-Dichlorophenyl)-1H-Pyrazole-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: 3HZ |
 |
Crystal Structure Of Rpa70N In Complex With 5-(4-((4-(5-Carboxyfuran-2-Yl)Phenylthioamido)Methyl)Phenyl)-1-(3,4-Dichlorophenyl)-1H-Pyrazole-3-Carboxylic Acid
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.28 Å Release Date: 2014-11-19 Classification: PROTEIN BINDING Ligands: 3J0 |
 |
Crystal Structure Of Rpa70N In Complex With A 3,4 Dichlorophenylalanine Atrip Derived Peptide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2014-02-26 Classification: PEPTIDE BINDING PROTEIN |

