Search Count: 83
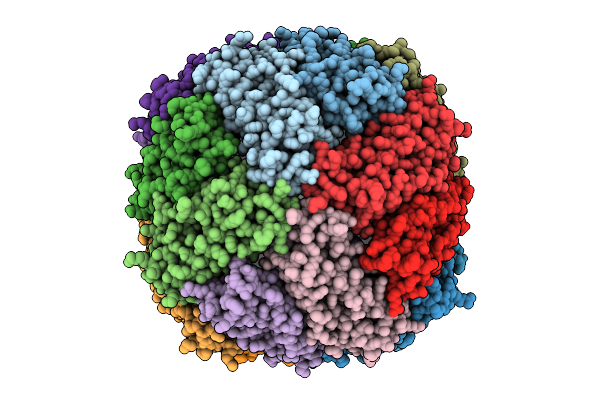 |
Organism: Magnetospirillum gryphiswaldense msr-1
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: METAL BINDING PROTEIN Ligands: HEM, FE |
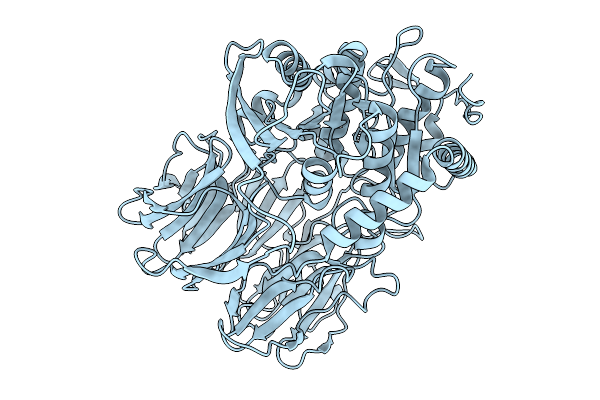 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE |
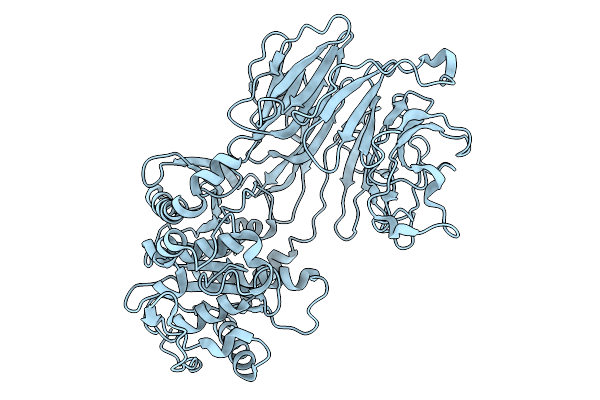 |
Organism: Trypanosoma cruzi
Method: ELECTRON MICROSCOPY Release Date: 2025-10-01 Classification: HYDROLASE |
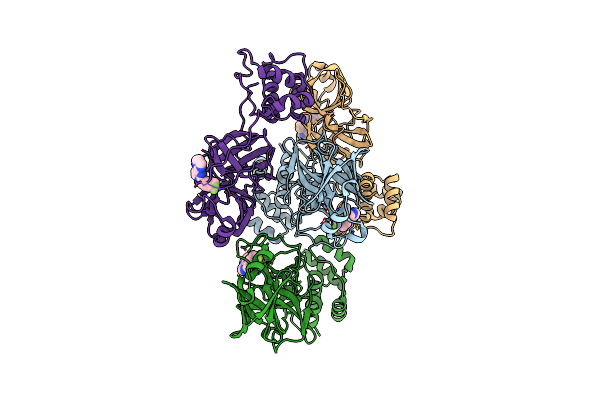 |
Organism: Human coronavirus oc43
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: A1AUY |
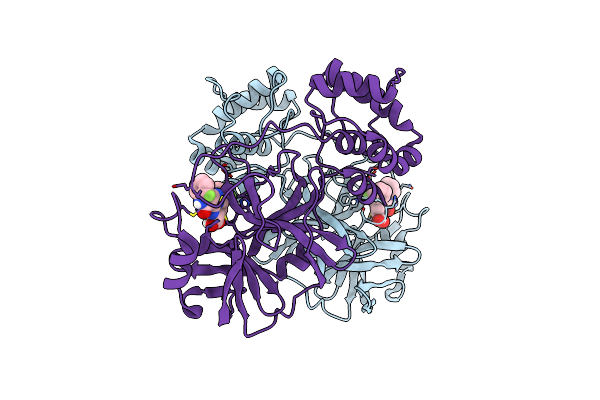 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2024-10-16 Classification: VIRAL PROTEIN Ligands: A1AUX |
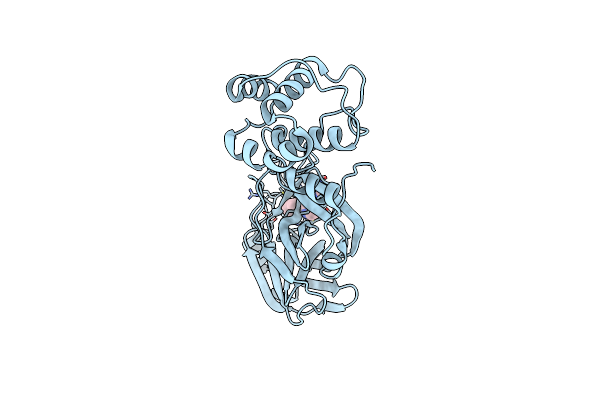 |
Co-Structure Of Main Protease Of Sars-Cov-2 (Covid-19) With Covalent Inhibitor
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-10-16 Classification: HYDROLASE Ligands: A1AU4 |
 |
Structures Of Small Molecules Bound To Rna Repeat Expansions That Cause Huntington'S Disease-Like 2 And Myotonic Dystrophy Type 1
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-08-07 Classification: RNA Ligands: MQC |
 |
Structures Of Small Molecules Bound To Rna Repeat Expansions That Cause Huntington'S Disease-Like 2 And Myotonic Dystrophy Type 1
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-08-07 Classification: RNA Ligands: A1AZM |
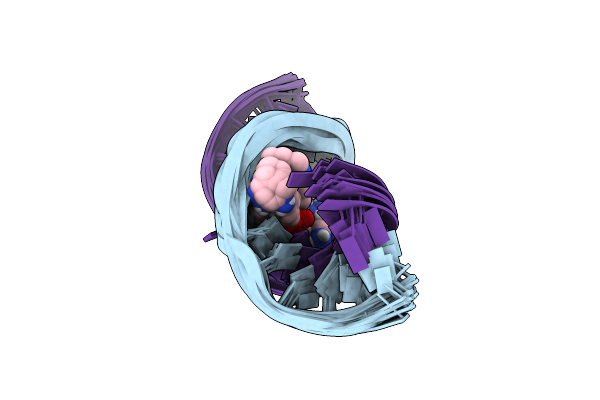 |
Structures Of Small Molecules Bound To Rna Repeat Expansions That Cause Huntington'S Disease-Like 2 And Myotonic Dystrophy Type 1
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2024-08-07 Classification: RNA Ligands: A1AZL |
 |
Structures Of Small Molecules Bound To Rna Repeat Expansions That Cause Huntington'S Disease-Like 2 And Myotonic Dystrophy Type 1
|
 |
Organism: Zika virus
Method: X-RAY DIFFRACTION Resolution:3.15 Å Release Date: 2023-09-06 Classification: RNA Ligands: MG |
 |
Cryo-Em Consensus Structure Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Minus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: MG |
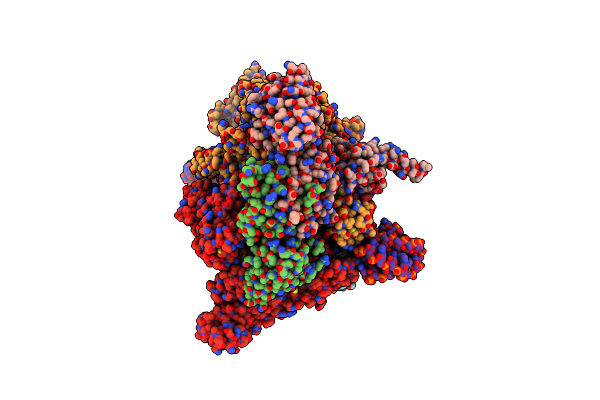 |
Cryo-Em Structure Of 3Dva Component 0 Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Minus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: MG |
 |
Cryo-Em Structure Of 3Dva Component 1 Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Minus Preq1 Ligand
Organism: Escherichia coli, Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: MG |
 |
Cryo-Em Structure Of 3Dva Component 2 Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Minus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.70 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: MG |
 |
Cryo-Em Consensus Structure Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Plus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.80 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: PRF, MG |
 |
Cryo-Em Structure Of 3Dva Component 0 Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Plus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:3.90 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: PRF, MG |
 |
Cryo-Em Structure Of 3Dva Component 1 Of Escherichia Coli Que-Pec (Paused Elongation Complex) Rna Polymerase Plus Preq1 Ligand
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:4.30 Å Release Date: 2023-06-21 Classification: TRANSCRIPTION Ligands: PRF, MG |
 |
 |

