Search Count: 40
 |
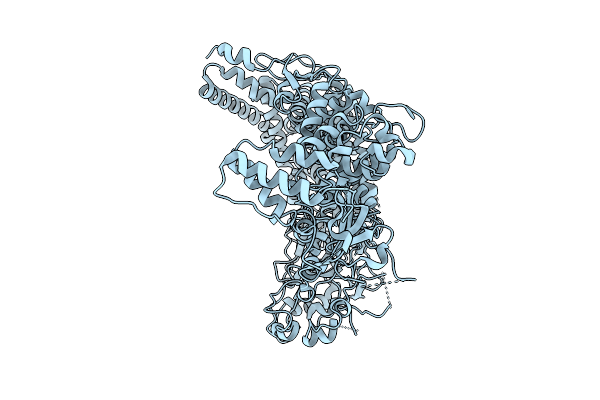
Cryo-Em Structure Of Essential Mycoplasma Pneumoniae Lipoprotein Mpn444 Homotrimer
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-09-17 Classification: UNKNOWN FUNCTION |
 |
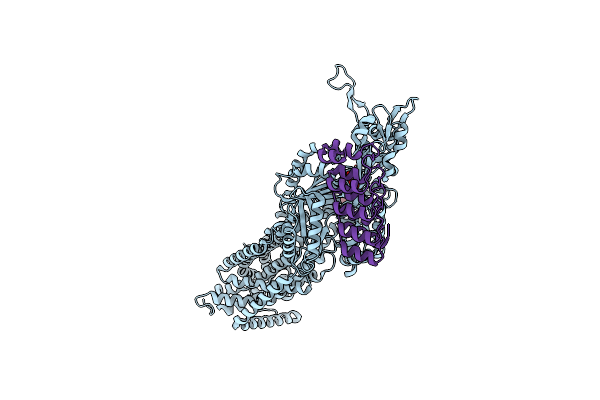
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2025-09-10 Classification: UNKNOWN FUNCTION |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN Ligands: MIY |
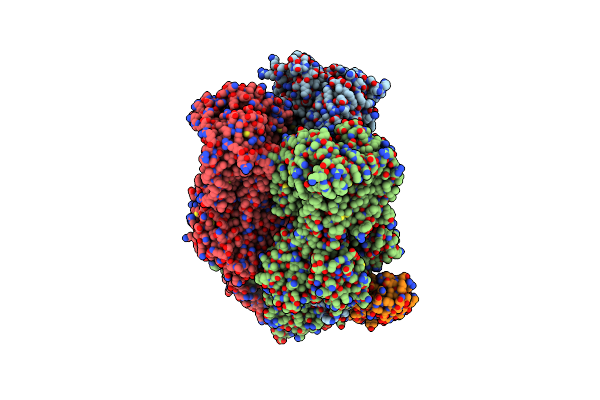 |
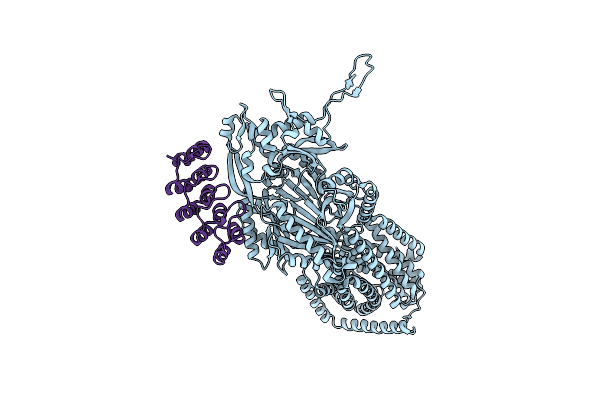
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
 |
Organism: Escherichia coli k-12, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-06-11 Classification: TRANSPORT PROTEIN |
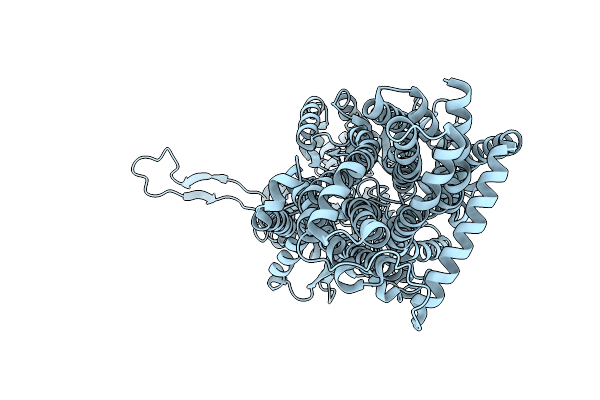 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
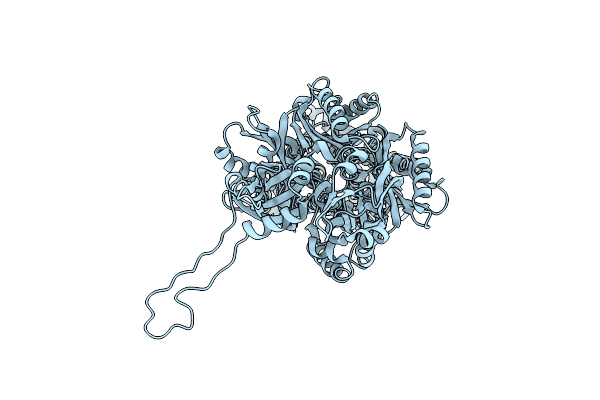 |
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
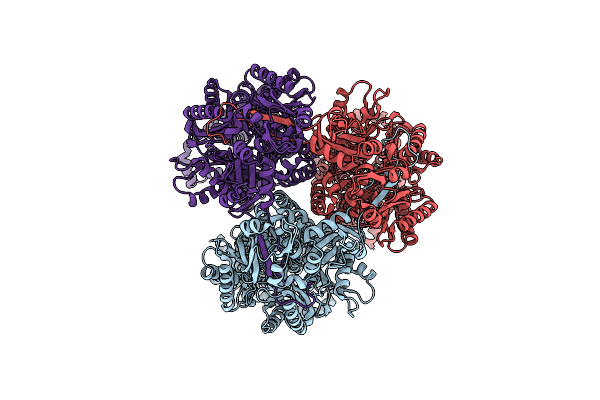 |
Single Particle Cryo-Em Structure Of The Multidrug Efflux Pump Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: ELECTRON MICROSCOPY Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
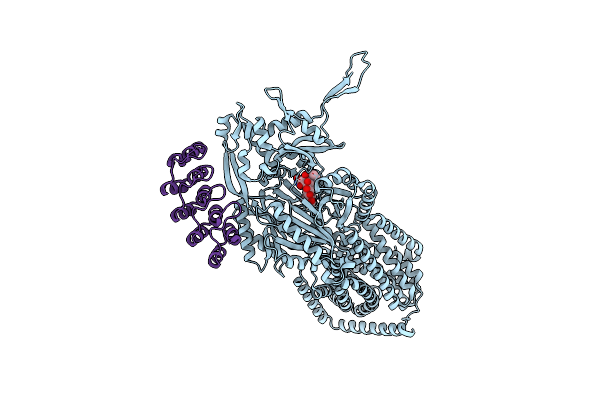 |
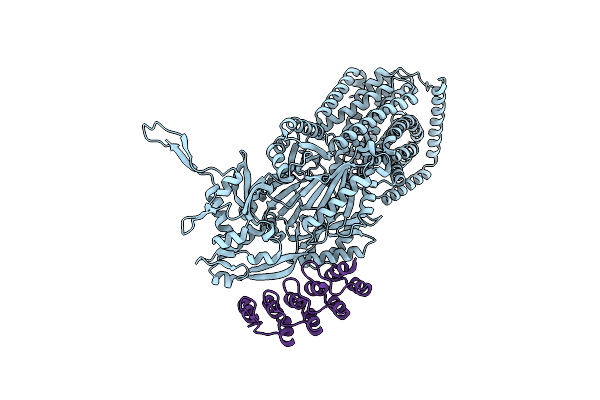
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN Ligands: MIY |
 |
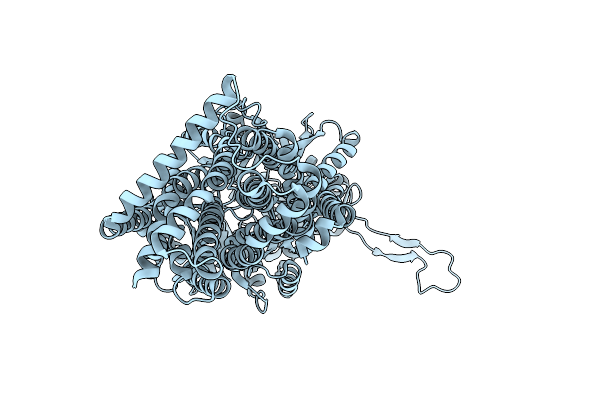
Organism: Synthetic construct, Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
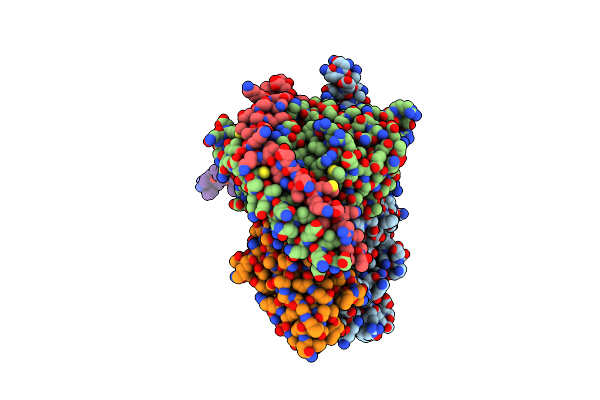
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: TRANSPORT PROTEIN |
 |
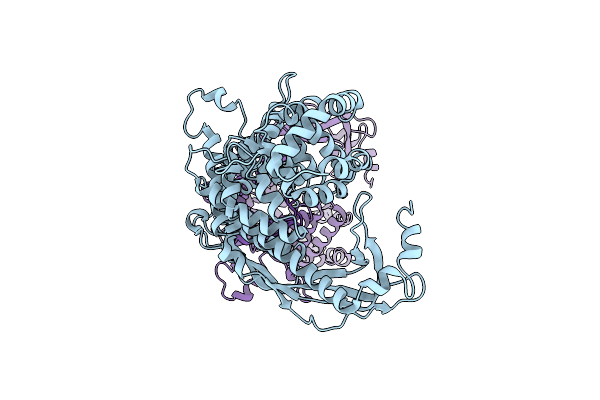
Cryo-Em Structure Of Cell-Free Synthesized Human Beta-1 Adrenergic Receptor Cotranslationally Inserted Into A Lipidic Nanodiscs
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-05-21 Classification: MEMBRANE PROTEIN Ligands: 5FW |
 |
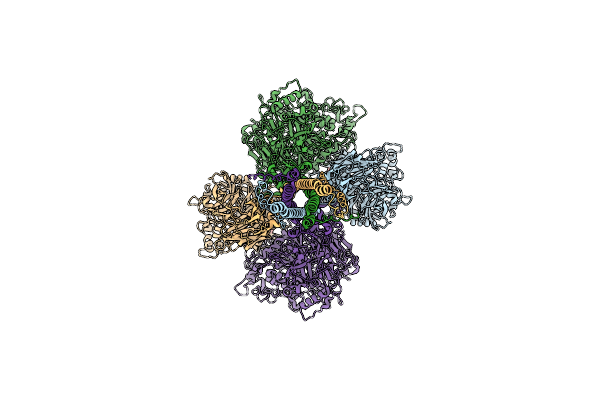
P116 Dimer In The Full State (Pdb Structure Of The Full-Length Ectodomain Truncated To Amino Acids 246-818)
Organism: Mycoplasmoides pneumoniae m129
Method: ELECTRON MICROSCOPY Release Date: 2024-05-29 Classification: LIPID BINDING PROTEIN |
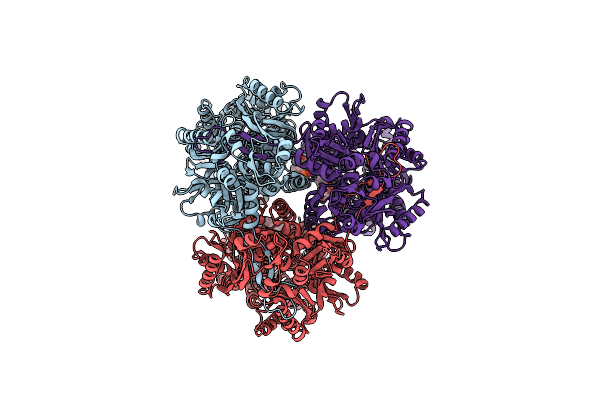 |
Single Particle Cryo-Em Co-Structure Of Klebsiella Pneumoniae Acrb With The Bdm91288 Efflux Pump Inhibitor At 2.97 Angstrom Resolution
Organism: Klebsiella pneumoniae subsp. pneumoniae dsm 30104 = jcm 1662 = nbrc 14940
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: WQW, WR6, PG8 |
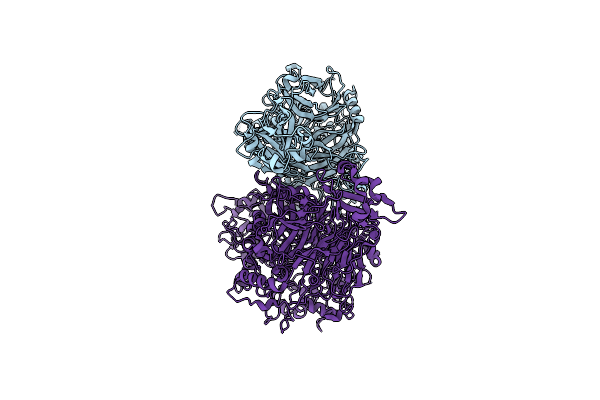 |
Single Particle Cryo-Em Of The P140-P110 Heterodimer Of Mycoplasma Genitalium At 3.3 Angstrom Resolution.
Organism: Mycoplasmoides genitalium g37
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: CELL ADHESION |
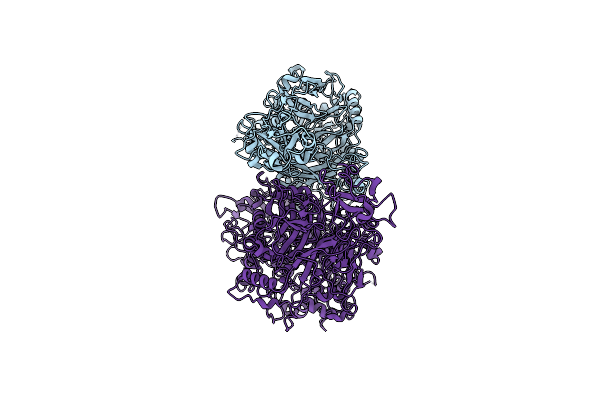 |
Single Particle Cryo-Em Of The P140-P110 Heterodimer With An Alternative Conformation In The P140 Stalk Of Mycoplasma Genitalium At A Resolution Of 3.7 Angstrom.
Organism: Mycoplasmoides genitalium g37
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: CELL ADHESION |
 |
Sub-Tomogram Average Of The Nap Adhesion Complex From The Human Pathogen Mycoplasma Genitalium At 11 Angstrom.
Organism: Mycoplasmoides genitalium g37
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: CELL ADHESION |
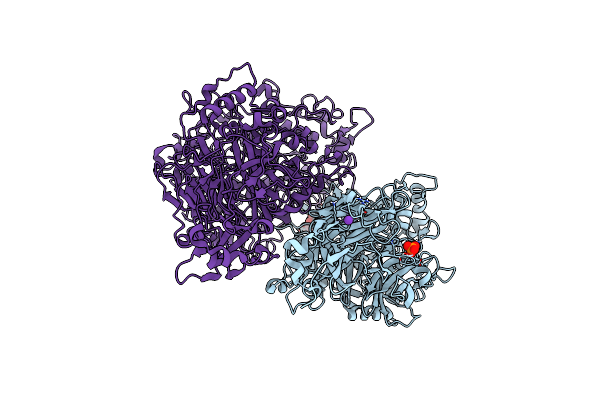 |
Sub-Tomogram Average Of The Open Conformation Of The Nap Adhesion Complex From The Human Pathogen Mycoplasma Genitalium.
Organism: Mycoplasmoides genitalium g37
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: CELL ADHESION Ligands: K, PO4 |
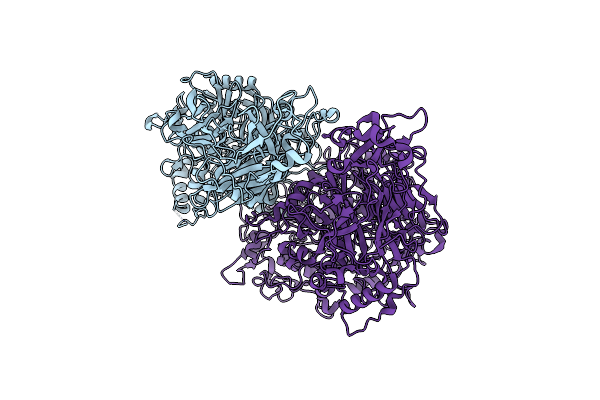 |
Sub-Tomogram Average Of The Closed Conformation Of The Nap Adhesion Complex From The Human Pathogen Mycoplasma Genitalium.
Organism: Mycoplasmoides genitalium g37
Method: ELECTRON MICROSCOPY Release Date: 2023-11-01 Classification: CELL ADHESION |
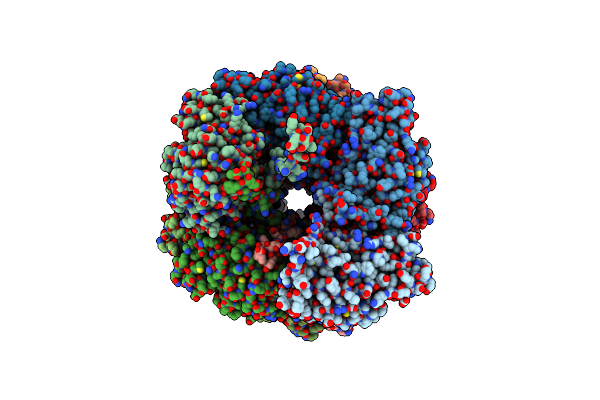 |
Organism: Bacillus pumilus
Method: ELECTRON MICROSCOPY Release Date: 2023-08-16 Classification: HYDROLASE |

