Search Count: 15
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Homocysteine Bound.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Methionine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM, EDO, GOL |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG, GOL |
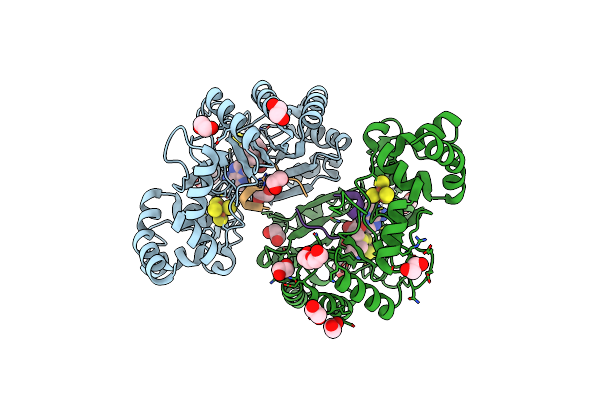 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 6 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
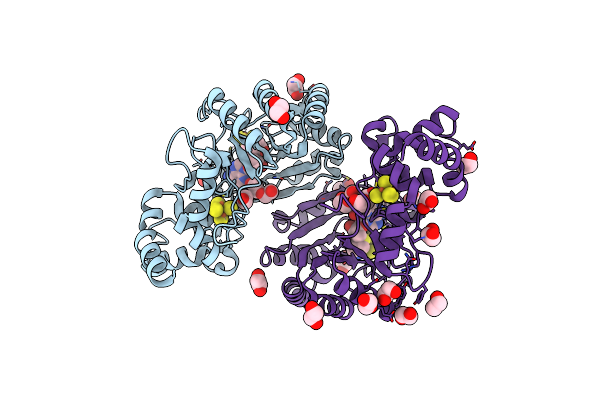 |
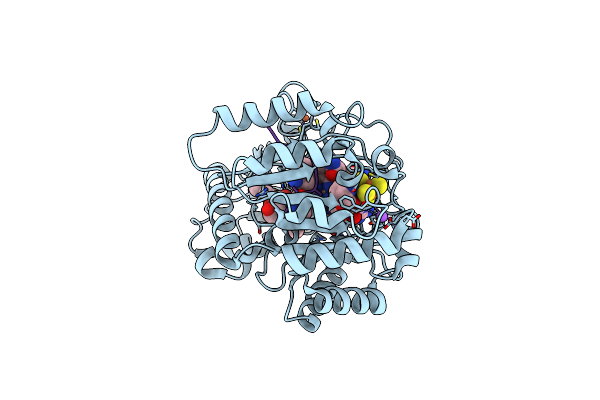
Crystal Structure Of Radical Sam Epimerase Epee D210A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Persulfurated Cysteine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
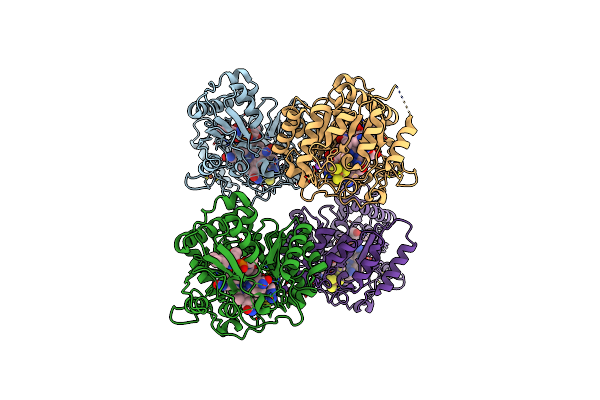
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, S-Adenosyl-L-Methionine, And Peptide Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, SAM, COB, NA |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA, PEG |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Adenosyl-L-Homocysteine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, SAH, COB, NA |
 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, As-Isolated (Protein Batch 1), Canonical C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, XCC, GOL, FES |
 |
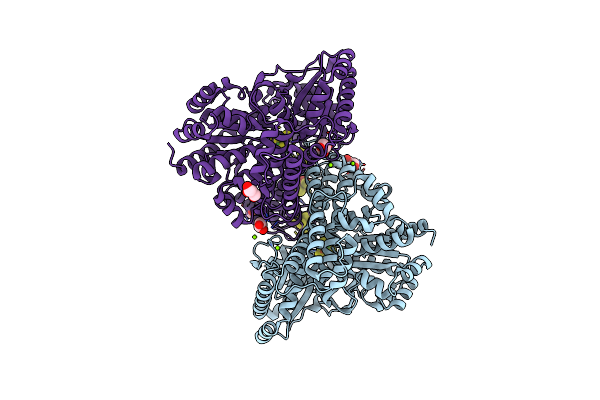
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, As-Isolated (Protein Batch 2), Oxidized C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, MG, XCC, CUV, FES, GOL, CL |
 |
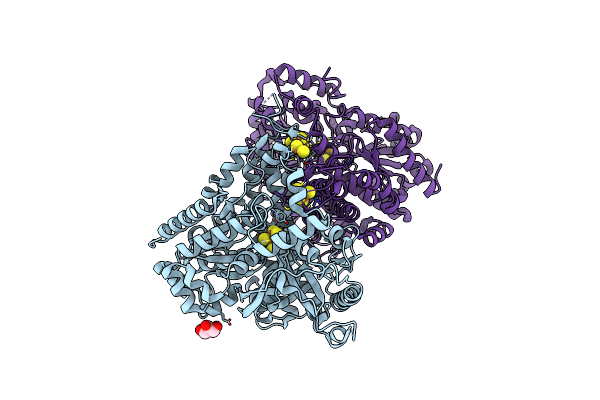
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced (Protein Batch 2), Canonical C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, XCC, GOL, MG, CL, FES |
 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase, Dithionite-Reduced Then Oxygen-Exposed (Protein Batch 2), Oxidized C-Cluster
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, CUV, GOL, FES |
 |
Crystal Structure Of Desulfovibrio Vulgaris Carbon Monoxide Dehydrogenase C301S Variant
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2018-10-03 Classification: OXIDOREDUCTASE Ligands: SF4, FES, XCC, FE, MG, GOL |

