Search Count: 147
 |
Organism: Penicillium rubens
Method: X-RAY DIFFRACTION Resolution:1.38 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, PEG |
 |
Organism: Penicillium rubens wisconsin 54-1255
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, A1ITD |
 |
Organism: Penicillium rubens wisconsin 54-1255
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, A1ITD |
 |
Organism: Penicillium rubens wisconsin 54-1255
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-03-19 Classification: OXIDOREDUCTASE Ligands: FAD, NAG, SCN |
 |
Organism: Penicillium steckii
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2025-03-05 Classification: OXIDOREDUCTASE Ligands: FMN, GOL, CL, CA |
 |
Organism: Rhodococcus jostii rha1
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2025-01-29 Classification: OXIDOREDUCTASE Ligands: FAD, B3P, MPD, MRD, CL |
 |
Crystal Structure Of Thermoanaerobacterales Bacterium Monoamine Oxidase In Complex With Putrescine
Organism: Thermoanaerobacterales bacterium
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2025-01-22 Classification: FLAVOPROTEIN Ligands: FAD, MG, PUT |
 |
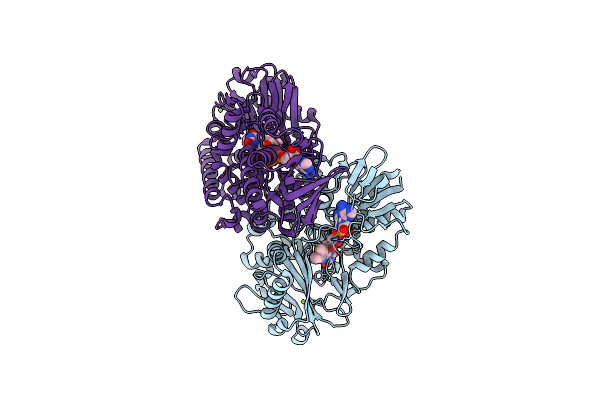
Crystal Structure Of Thermoanaerobacterales Bacterium Monoamine Oxidase In Complex With Spermidine And Its Oxidation Products
Organism: Thermoanaerobacterales bacterium
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-22 Classification: FLAVOPROTEIN Ligands: FAD, GOL, 13D, 4HA, MG, SPD |
 |
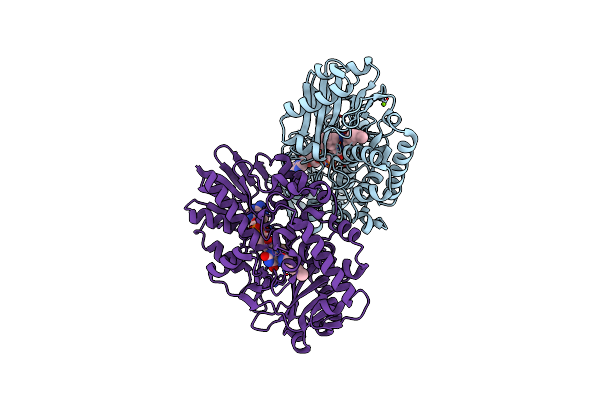
Crystal Structure Of Thermoanaerobacterales Bacterium Monoamine Oxidase In Complex With Benzylamine
Organism: Thermoanaerobacterales bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-22 Classification: FLAVOPROTEIN Ligands: FAD, MG, ABN |
 |
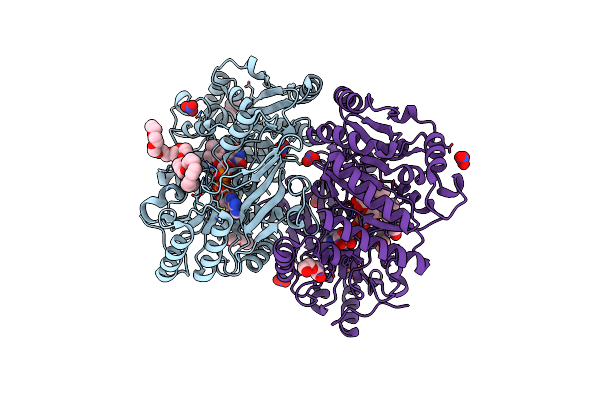
Crystal Structure Of Thermoanaerobacterales Bacterium Monoamine Oxidase In Complex With N-Heptylamine
Organism: Thermoanaerobacterales bacterium
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-22 Classification: FLAVOPROTEIN Ligands: FAD, MG, A1ISO |
 |
Vanillyl-Alcohol Dehydrogenase From Marinicaulis Flavus: P151L Mutant Bound To Eugenol
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, 1PE, PE4, EOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
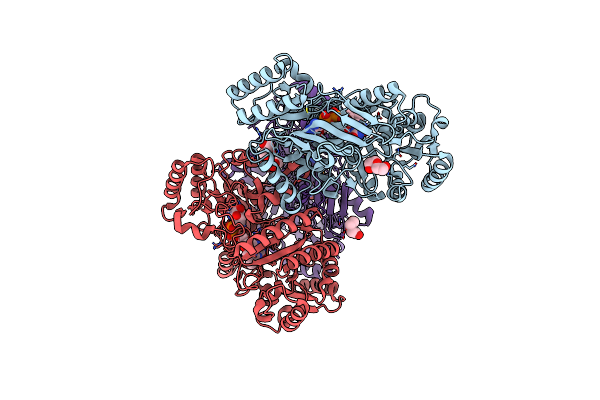
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, GOL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, GOL, PEG |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, CL |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, NO3, PEG, SO4 |
 |
Organism: Marinicaulis flavus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2025-01-15 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PEG |
 |
Vanillyl Alcohol Oxidase From Novosphingobium Sp In Complex With Vanillyl Alcohol
Organism: Novosphingobium sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-01-15 Classification: HYDROLASE Ligands: FAD, GOL, V55 |
 |
Vanillyl Alcohol Oxidase From Novosphingobium Sp: T181D Mutant In Complex With Vanillin
Organism: Novosphingobium sp. 01wb02.4-10
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-01-15 Classification: FLAVOPROTEIN Ligands: FAD, V55, GOL, PEG |

