Search Count: 7
 |
Solution Structure Of Lysm The Peptidoglycan Binding Domain Of Autolysin Atla From Enterococcus Faecalis
Organism: Enterococcus faecalis
Method: SOLUTION NMR Release Date: 2014-06-18 Classification: HYDROLASE |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-05-25 Classification: HYDROLASE |
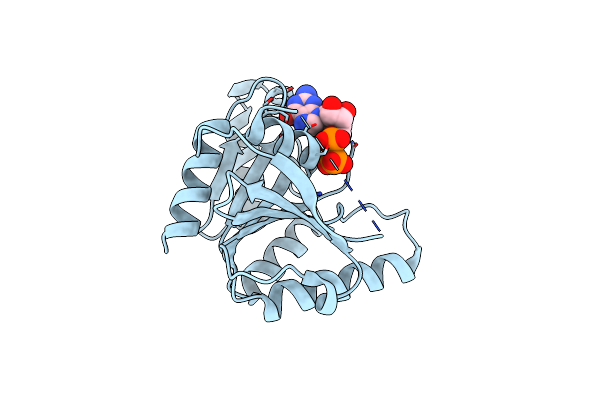 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-05-25 Classification: HYDROLASE Ligands: GDP |
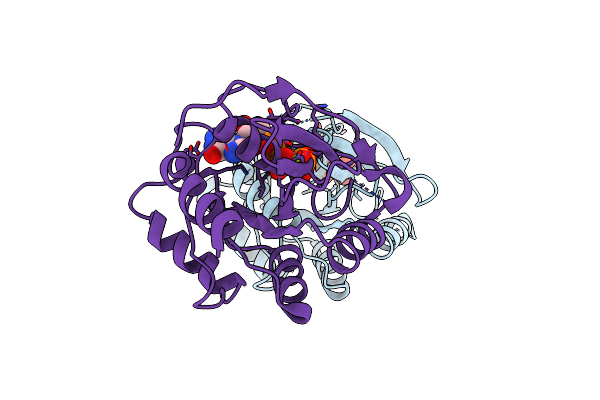 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2004-05-25 Classification: HYDROLASE Ligands: MG, GTP |
 |
The 2.3 Angstrom Resolution Structure Of Bacillus Subtilis Luxs/Homocysteine Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2001-10-24 Classification: SIGNALING PROTEIN Ligands: ZN, HCS |
 |
The 2.2 Angstrom Resolution Structure Of Bacillus Subtilis Luxs/Ribosilhomocysteine Complex
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2001-10-24 Classification: SIGNALING PROTEIN Ligands: ZN, SO4, RHC, HCS |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2001-06-06 Classification: SIGNALING PROTEIN Ligands: ZN |

