Search Count: 40
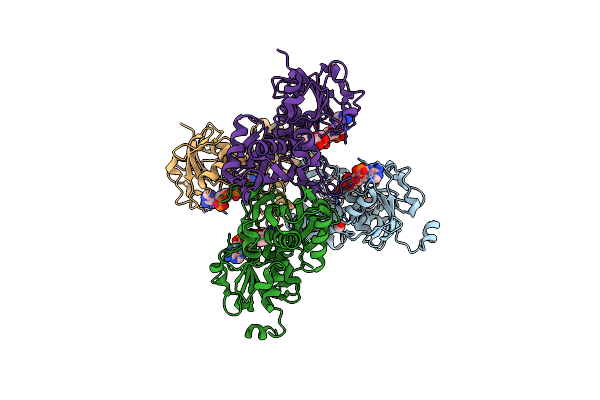 |
Glyceraldehyde 3-Phosphate Dehydrogenase (Gapa) From Helicobacter Pylori In Complex With Napd (Holo)
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: NAP, MOH |
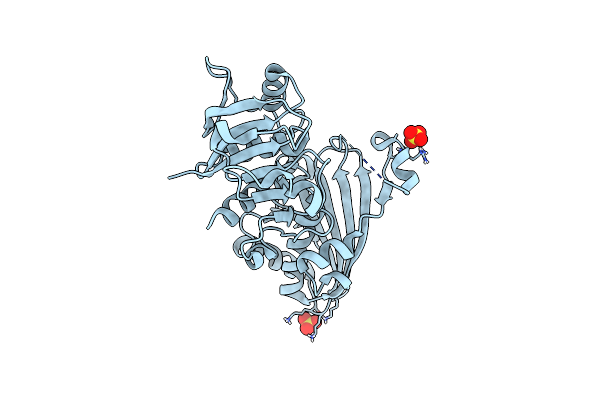 |
Glyceraldehyde 3-Phosphate Dehydrogenase A (Gapdha) Apoenzyme, From Helicobacter Pylori
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: SO4 |
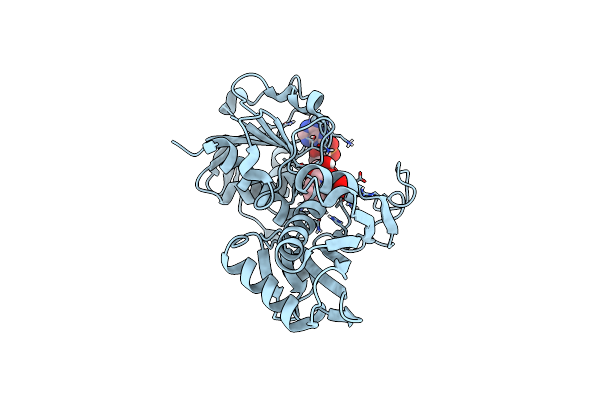 |
Glyceraldehyde 3-Phosphate Dehydrogenase A (Gapdha) Nadp Holoenzyme, From Helicobacter Pylori, With Active Site Cysteine Oxidised To Sulfenic Acid
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-25 Classification: OXIDOREDUCTASE Ligands: NAP, SO4 |
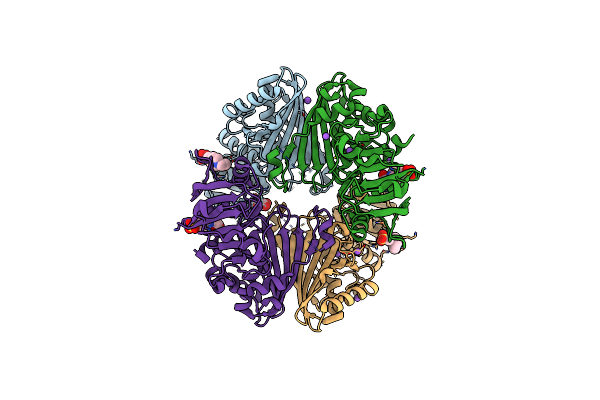 |
Apo Glyceraldehyde 3-Phosphate Dehydrogenase (Gapa) From Helicobacter Pylori
Organism: Helicobacter pylori 26695
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: EPE, GOL, NA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-01-11 Classification: PROTEIN BINDING/Immune System |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex In The Ligand-Free Form At 303 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2022-11-09 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Aifs In The Ligand-Free Form At 100 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Aifs Bound To Maleic Acid At 100 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: MAE, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2022-11-02 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase Bound To Maleate At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, SO4 |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfit Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfiy In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: SO4, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfiy Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Hex Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfcs In The Ligand-Free Form At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: SO4, PLP |
 |
Crystal Structure Of E. Coli Aspartate Aminotransferase Mutant Vfcs Bound To Maleic Acid At 278 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, MAE |
 |
Crystal Structure Of Wild-Type E. Coli Aspartate Aminotransferase In The Ligand-Free Form At 303 K
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2022-10-05 Classification: TRANSFERASE Ligands: PLP, SO4 |

