Search Count: 248
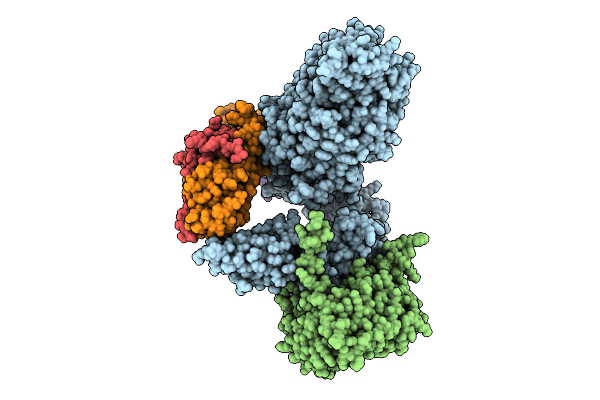 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
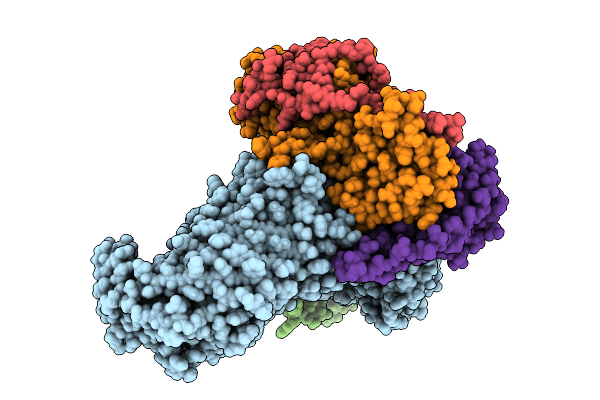 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
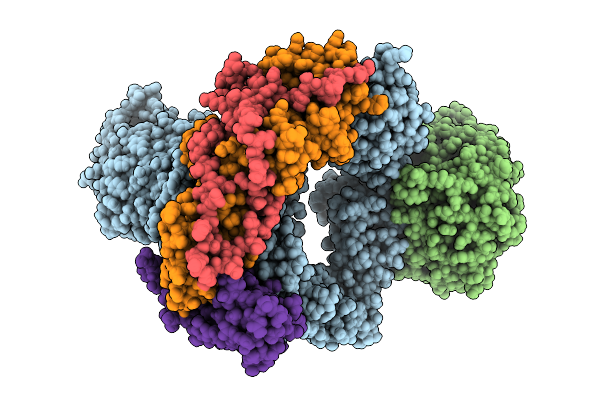 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-10-22 Classification: MEMBRANE PROTEIN |
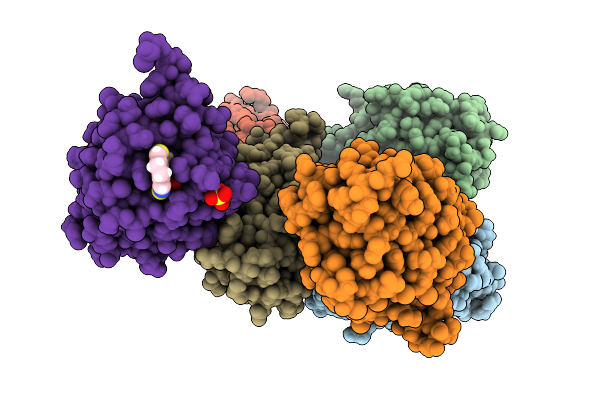 |
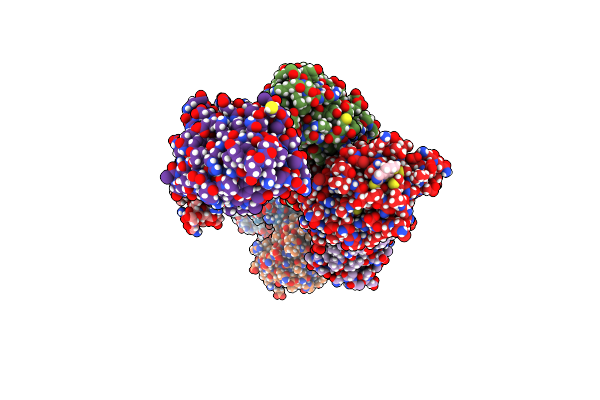
Crystal Structure Of An Mkp5 Mutant, Y435F, In Complex With An Allosteric Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: HYDROLASE/INHIBITOR Ligands: CJA, SO4 |
 |
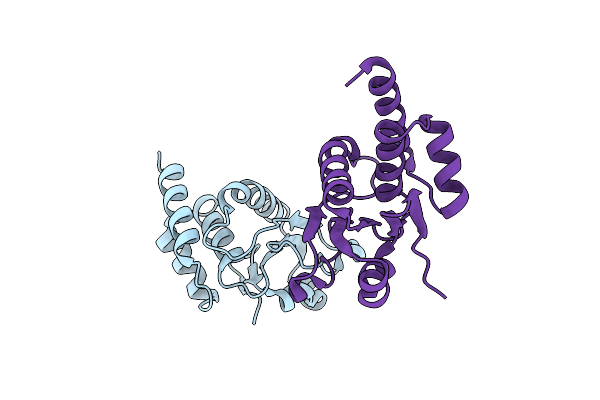
Crystal Structure Of An Mkp5 Mutant, Y435W, In Complex With An Allosteric Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-05-28 Classification: HYDROLASE/INHIBITOR Ligands: CJA |
 |
 |
Cryo-Em Structure Of Kbtbd4 Wt-Hdac2 2:1 Complex Mediated By Molecular Glue Um171
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: LIGASE Ligands: ZN, A1ACV |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: LIGASE Ligands: ZN |
 |
Cryo-Em Structure Of Kbtbd4 Wt-Hdac2 2:2 Complex Mediated By Molecular Glue Um171
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: LIGASE Ligands: A1ACV, ZN |
 |
Cryo-Em Structure Of Kbtbd4 Wt-Hdac2-Corest1 2:1:1 Complex Mediated By Molecular Glue Um171
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-04-09 Classification: LIGASE Ligands: A1ACV, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
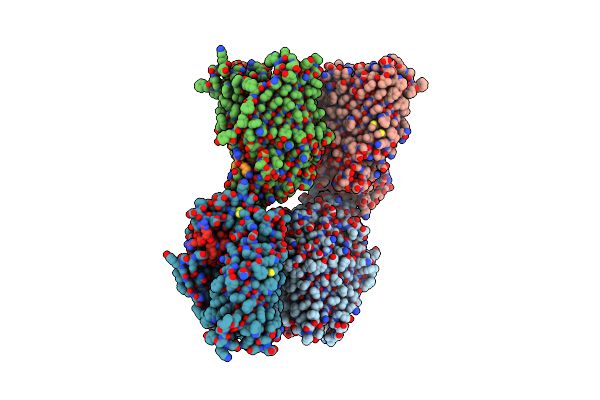
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-05 Classification: PROTEIN FIBRIL |
 |
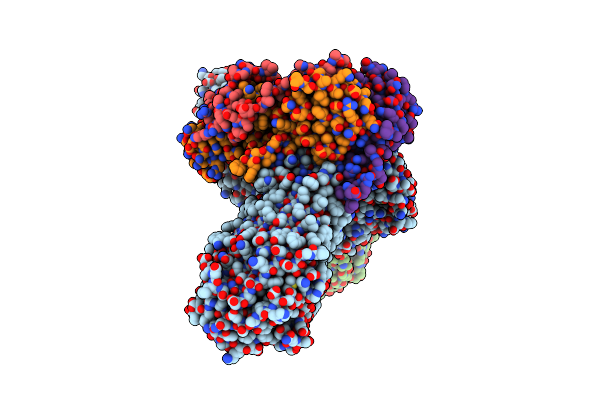
Crystal Structure Of Bama In Complex With The Ptb2 Open-State Inhibitor (Anisotropic Data Set)
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2025-01-15 Classification: MEMBRANE PROTEIN |
 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: MEMBRANE PROTEIN |
 |
Cryoem Structure Of Bam In Complex With The Ptb1 Closed-State Inhibitor (In Ddm Detergent)
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: MEMBRANE PROTEIN Ligands: ZN |

