Search Count: 54
 |
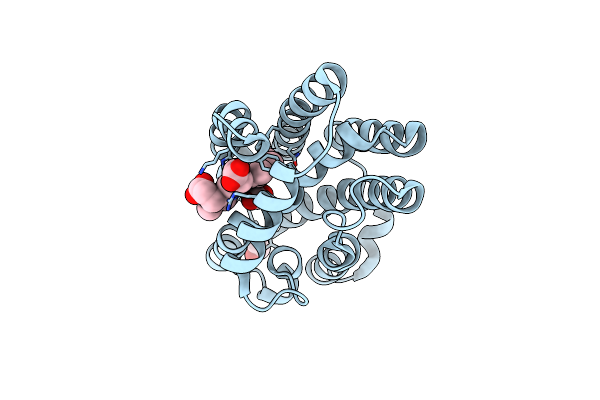
Crystal Structure Of Heme-Oxygenase From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.15 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, GOL, EDO, CIT, CL |
 |
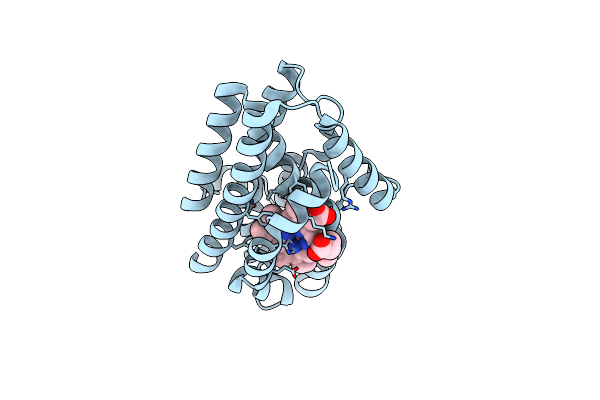
Crystal Structure Of Heme-Oxygenase From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii)) Flash-Cooled Under Co2 Pressure
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, EDO, CL, CO2 |
 |
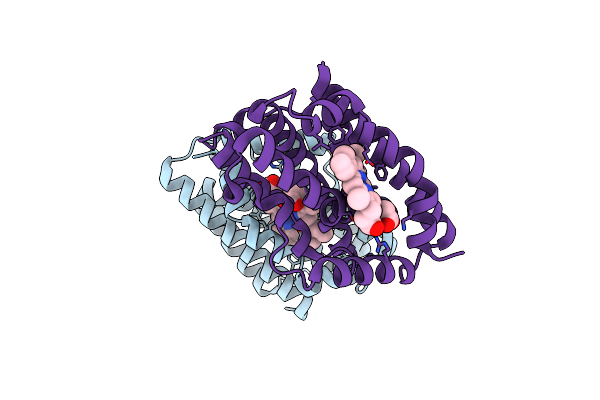
Crystal Structure Of Heme-Oxygenase Mutant G139A From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH, CL |
 |
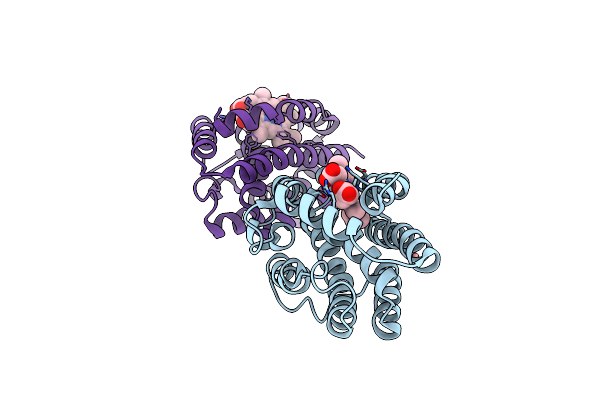
Crystal Structure Of Heme-Oxygenase Mutant H20C From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: COH |
 |
Crystal Structure Of Heme-Oxygenase Mutant I143K From Corynebacterium Diphtheriae Complexed With Cobalt-Porphyrine (Humo-Co(Iii))
Organism: Corynebacterium diphtheriae
Method: X-RAY DIFFRACTION Resolution:2.84 Å Release Date: 2024-10-16 Classification: OXIDOREDUCTASE Ligands: CO, COH, CL |
 |
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SF4, IOD, CL |
 |
Organism: Methanococcus maripaludis s2
Method: X-RAY DIFFRACTION Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: ANP, BME, SO4, GOL |
 |
Organism: Methanococcus maripaludis
Method: X-RAY DIFFRACTION Release Date: 2024-01-31 Classification: TRANSFERASE Ligands: SF4, AMP, CL |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2023-04-05 Classification: FLUORESCENT PROTEIN Ligands: PEG, MES, EDO, GOL |
 |
Organism: Discosoma sp.
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2023-04-05 Classification: FLUORESCENT PROTEIN Ligands: GOL, MES, PEG, EDO |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-07-21 Classification: TRANSFERASE Ligands: HUF, PEG, PO4, FDA, PG4 |
 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2021-07-21 Classification: TRANSFERASE Ligands: PGE, HUF, FDA, PEG |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Solved By Fe-Sad Phasing
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, PEG |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, CL |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, PGE, CL |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Soaked With 4-Thiouracil (12.65 Kev, 7.125 Kev (Fe-Sad) And 6.5 Kev (S-Sad) Data)
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.33 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, H2S, EDO, PEG |
 |
[4Fe-4S]-Dependent Thiouracil Desulfidase Tuds (Duf523Vcz) Soaked With 4-Thiouracil (S-Sad Data)
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2020-09-30 Classification: TRANSFERASE Ligands: SF4, EDO, H2S, PEG |
 |
Structure Of The Trna-Monooxygenase Enzyme Miae Frozen Under 140 Bar Of Krypton Using The Soak And Freeze Methodology
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-09-02 Classification: OXIDOREDUCTASE Ligands: FE, CL, CA, TRS, PEG, PG4, KR, EDO |
 |
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2020-09-02 Classification: OXIDOREDUCTASE Ligands: FE, CL, CA, TRS, PEG, PG4, PGE, PG6 |
 |
Structure Of The Trna-Monooxygenase Enzyme Miae Frozen Under 2000 Bar Using The High Pressure Freezing Method
Organism: Pseudomonas putida kt2440
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-09-02 Classification: OXIDOREDUCTASE Ligands: FE, CL, CA, TRS, PG4, AR, PEG |

