Search Count: 16
 |
Organism: Salmonella typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Structure Of The Rbm3/Collar Region Of The Salmonella Flagella Ms-Ring Protein Flif With 33-Fold Symmetry Applied
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Structure Of The Rbm2Inner Region Of The Salmonella Flagella Ms-Ring Protein Flif With 21-Fold Symmetry Applied.
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Structure Of The Rbm3/Collar Region Of The Salmonella Flagella Ms-Ring Protein Flif With 34-Fold Symmetry Applied
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Structure Of The Rbm2 Inner Ring Of Salmonella Flagella Ms-Ring Protein Flif With 22-Fold Symmetry Applied
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Structure Of The Rbm3/Collar Region Of The Salmonella Flagella Ms-Ring Protein Flif With 32-Fold Symmetry Applied
Organism: Salmonella enterica subsp. enterica serovar typhimurium
Method: ELECTRON MICROSCOPY Release Date: 2020-03-18 Classification: MOTOR PROTEIN |
 |
Organism: Klebsiella pneumoniae subsp. rhinoscleromatis sb3432
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2017-12-13 Classification: HYDROLASE Ligands: GNP, NI |
 |
Organism: Oryza sativa japonica group
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2015-08-26 Classification: LIGASE Ligands: K |
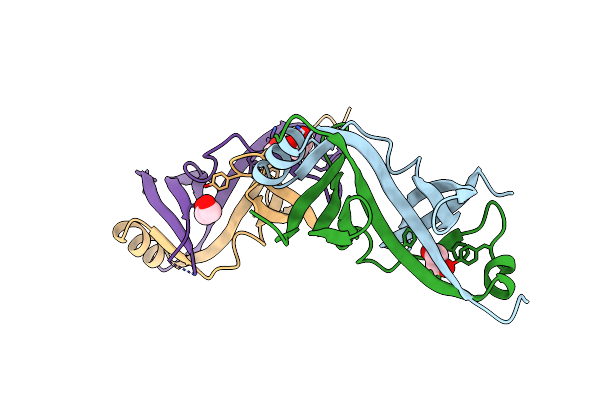 |
Organism: Shigella flexneri
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2015-06-24 Classification: PROTEIN TRANSPORT Ligands: EDO |
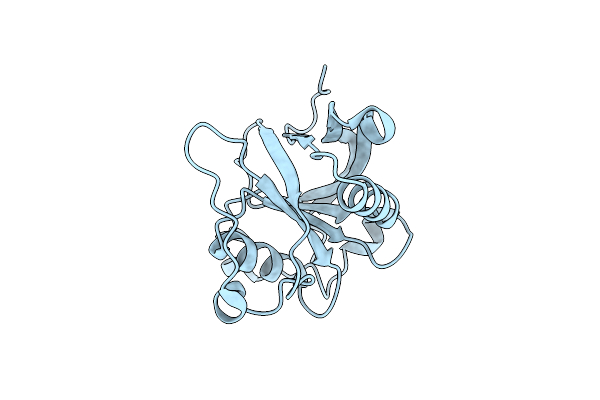 |
Crystal Structure Of Protease-Associated Domain Of Arabidopsis Vacuolar Sorting Receptor 1
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2014-12-10 Classification: PROTEIN TRANSPORT Ligands: IOD |
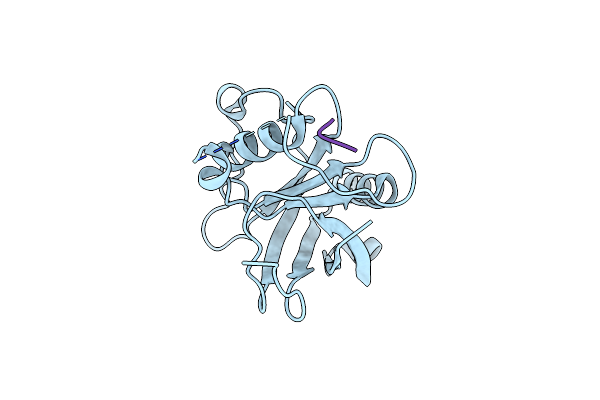 |
Crystal Structure Of Protease-Associated Domain Of Arabidopsis Vsr1 In Complex With Aleurain Peptide
Organism: Arabidopsis thaliana, Hordeum vulgare
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2014-12-10 Classification: PROTEIN TRANSPORT |
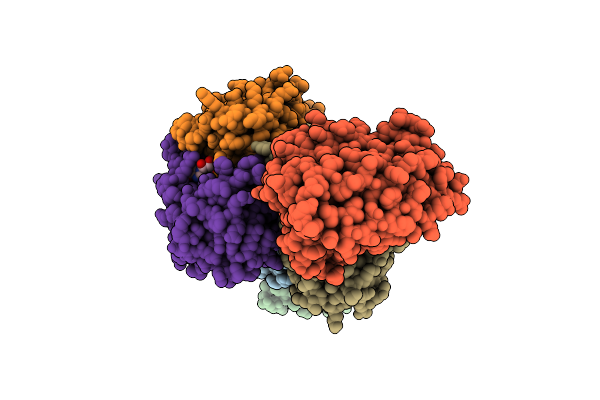 |
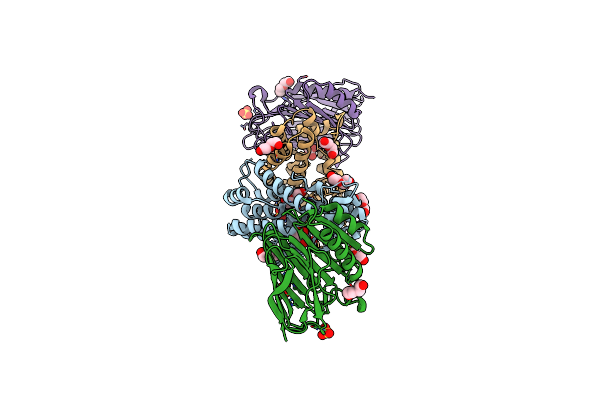
Crystal Structure Of Helicobacter Pylori Urease Accessory Protein Uref/H/G Complex
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-10-16 Classification: METAL BINDING PROTEIN Ligands: GDP |
 |
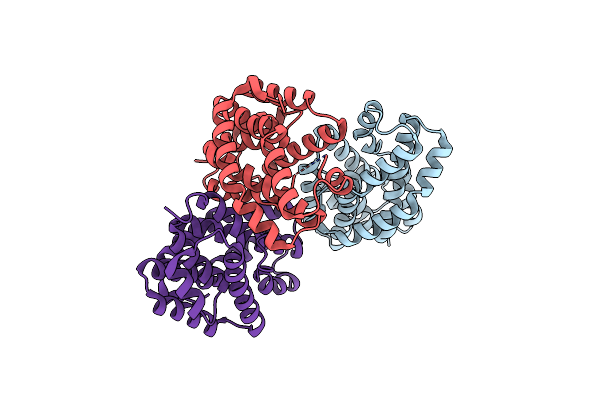
Crystal Structure Of Helicobacter Pylori Urease Accessory Protein Uref/H Complex
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-11-02 Classification: CHAPERONE Ligands: PEG, GOL, SO4 |
 |
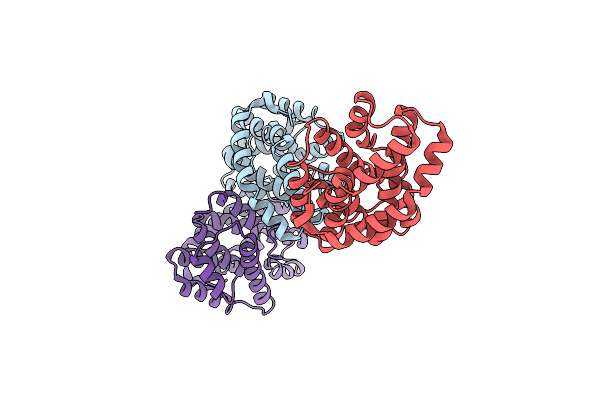
Native Crystal Structure Of Helicobacter Pylori Urease Accessory Protein Uref
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-08-18 Classification: METAL BINDING PROTEIN |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-05-26 Classification: METAL BINDING PROTEIN |

