Search Count: 4
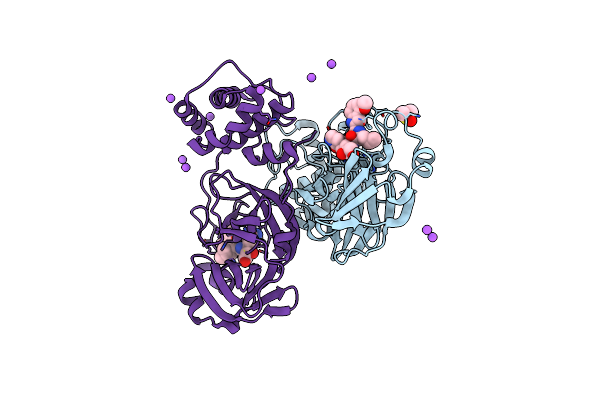 |
Crystal Structure Of Sars Cov-2 Mpro Mutant L141R With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN Ligands: DMS, V2M, NA |
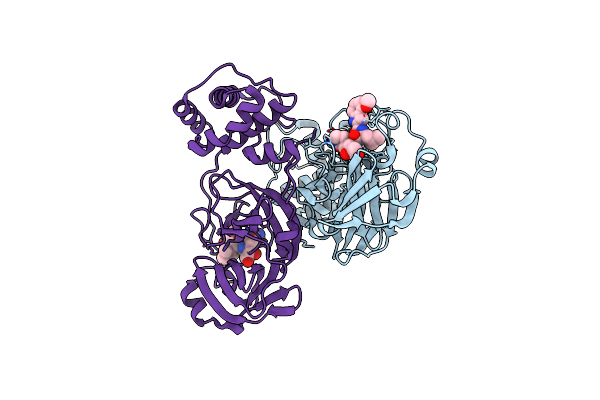 |
Crystal Structure Of Sars Cov-2 Mpro Mutant N142P With Pfizer Intravenous Inhibitor Pf-00835231
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: V2M |
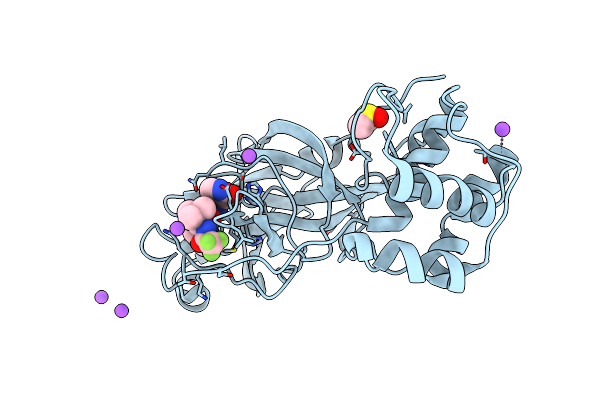 |
Crystal Structure Of Sars Cov-2 Mpro Mutant L50F With Nirmatrelvir Captured In Two Conformational States
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2024-02-07 Classification: VIRAL PROTEIN, HYDROLASE/INHIBITOR Ligands: DMS, 4WI, NA, CL |
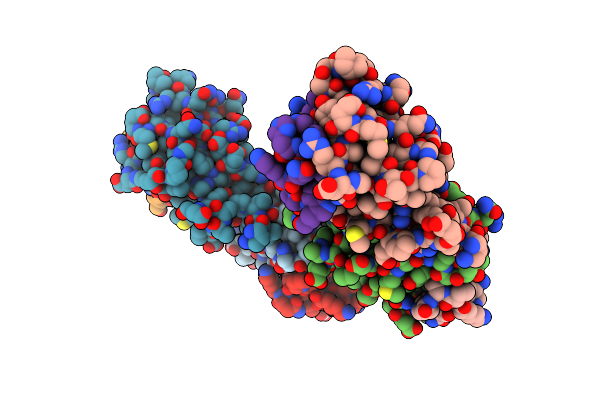 |
Versatile Modes Of Peptide Recognition By The Aaa+ Adaptor Protein Sspb- The Crystal Structure Of A Sspb-Rsea Complex
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2005-05-17 Classification: PROTEIN BINDING |

